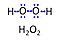- Development of a consistent nomenclature in mitochondrial and chloroplast physiology, in the spirit of Gentle Science
- high-resolution terminology - matching measurements at high-resolution
Introduction to MitoPedia
- The term MitoPedia expresses the link to Wikipedia. We use the MediaWiki software for generating a bottom-up database and glossaries. The 'Bioblast community' of mitochondrial experts is cordially invited to active participation - a vision that may develop further in the future. To make this possible, the Oroboros-team continues to work on the Bioblast website as an information synthase - in the spirit of Gentle Science and as a branch of the Oroboros Ecosystem. ~ Gnaiger Erich 09:20, 18 May 2014 (CEST)
SI, IUPAC and MitoEAGLE recommendations
- 'The International System of Units, the SI, has been used around the world as the preferred system of units, the basic language for science, technology, industry and trade since it was established in 1960'[6]. IUPAC guidelines are followed for general terms of physical chemistry[7],[8],[9], extended by concepts of mitochondrial physiology and nonequilibrium thermodynamics[4],[10],[11].
- According to BEC guidelines, 'manuscripts must adhere to SI units and IUPAC recommendations. MitoEAGLE recommendations on terms and symbols are to be implemented'. Harmonization of terms helps to link to the general literature.
MitoPedia: Concepts
MitoPedia: MiP and biochemistry
MitoPedia: Methods
MitoPedia: O2k and high-resolution respirometry
MitoPedia: BEC, preprints, and history
- » MitoPedia: BEC - Bioenergetics Communications
- » MitoPedia: Preprints - MitoFit Preprints
- » Mitochondria and bioblasts: Made history
References
- ↑ Cohen ER, Cvitas T, Frey JG, Holmström B, Kuchitsu K, Marquardt R, Mills I, Pavese F, Quack M, Stohner J, Strauss HL, Takami M, Thor HL (2008) Quantities, Units and Symbols in Physical Chemistry, IUPAC Green Book, 3rd Edition, 2nd Printing, IUPAC & RSC Publishing, Cambridge. - »Bioblast link«
- ↑ International Union of Biochemistry and Molecular Biology: Recommendations for terminology and databases for biochemical thermodynamics. - »Open Access«
- ↑ International Union of Biochemistry (1981) Symbolism and terminology in enzyme kinetics. - »Open Access«
- ↑ 4.0 4.1 4.2 4.3 Gnaiger E (2020) Mitochondrial pathways and respiratory control. An introduction to OXPHOS analysis. 5th ed. Bioenerg Commun 2020.2. https://doi.org/10.26124/bec:2020-0002 - »Bioblast link«
- ↑ Gnaiger E (1993) Nonequilibrium thermodynamics of energy transformations. Pure Appl Chem 65:1983-2002. - »Bioblast link«
- ↑ Bureau International des Poids et Mesures (2019) The International System of Units (SI). 9th edition:117-216. ISBN 978-92-822-2272-0
- ↑ Cohen ER, Cvitas T, Frey JG, Holmström B, Kuchitsu K, Marquardt R, Mills I, Pavese F, Quack M, Stohner J, Strauss HL, Takami M, Thor HL (2008) Quantities, Units and Symbols in Physical Chemistry, IUPAC Green Book, 3rd Edition, 2nd Printing, IUPAC & RSC Publishing, Cambridge. - »Bioblast link«
- ↑ International Union of Biochemistry and Molecular Biology: Recommendations for terminology and databases for biochemical thermodynamics. - »Open Access«
- ↑ International Union of Biochemistry (1981) Symbolism and terminology in enzyme kinetics. - »Open Access«
- ↑ Gnaiger Erich et al ― MitoEAGLE Task Group (2020) Mitochondrial physiology. Bioenerg Commun 2020.1. doi:10.26124/bec:2020-0001.v1
- ↑ Gnaiger E (1993) Nonequilibrium thermodynamics of energy transformations. Pure Appl Chem 65:1983-2002. - »Bioblast link«
- ↑ International Union of Biochemistry (1991) Nomenclature of electron-transfer proteins. Biochim Biophys Acta 1060. - »Open Access«
MitoPedia terms and abbreviations
| Term | Abbreviation | Description |
|---|---|---|
| % | % | The symbol % indicates 'per cent' (per hundred). {Quote} The internationally recognized symbol % (per cent) may be used with the SI. When it is used, a space separates the number and the symbol %. {end of Quote}. |
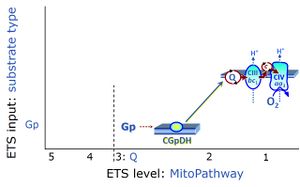
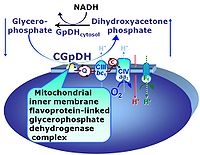
| 1OctM;2D;3PG;4S;5U;6Rot- | FNS(Oct,PGM) | 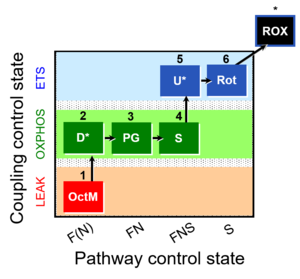 |
| 1PGM;2D;3S;4Rot;5U- | NS(PGM) | 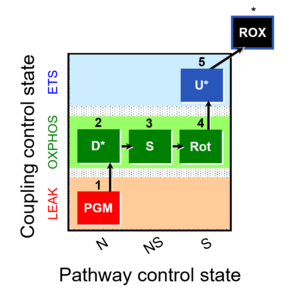 |
| 1PGM;2D;3U;4S;5Rot- | NS(PGM) | 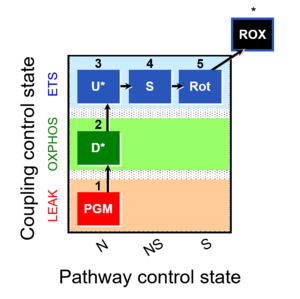 |
| 1PM;2D;3G;4U;5S;6Rot- | NS(PGM) | 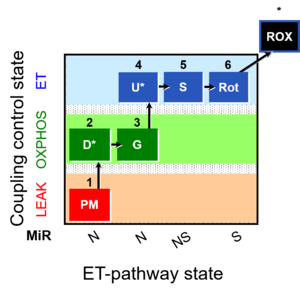 |
| 2-Deoxyglucose | 2-DG | 2-Deoxyglucose, also known as 2-deoxy-D-glucose is a glucose derivative that has the 2-hydroxyl group replaced by hydrogen. It competitively inhibits glycolysis by blocking hexokinase and phosphohexoseisomerase. |
| 2-Hydroxyglutarate | 2HG | Reduction of oxoglutarate (2OG or alpha-ketoglutarate) to 2-hydroxyglutarate (2HG) is driven by NADPH. 2HG is also formed in side reactions of lactate dehydrogenase and malate dehydrogenase. Millimolar 2HG concentrations are found in some cancer cells compared to , whereas side activities of lactate and malate dehydrogenase form submillimolar s-2-hydroxyglutarate (s-2HG). However, even wild-type IDH1 and IDH2, notably under shifts toward reductive carboxylation glutaminolysis or changes in other enzymes, lead to “intermediate” 0.01–0.1 mM 2HG levels, for example, in breast carcinoma compared with nanomolar concentrations in benign cells. 2HG is considered an important player in reprogramming metabolism of cancer cells. |
| 2-mercaptoacetate | 2-mercaptoacetate is an inhibitor of medium-chain acyl-CoA dehydrogenase, MCAD, the rate-limiting enzyme of octanoylcarnitine oxidation. 2-mercaptoacetate has been used as an inhibitor of fatty acid oxidation (F-pathway control state). In permeabilized rat soleus muscle fibers, pre-incubation with 1 mM 2-mercaptoacetate for 45 min resulted in 58% inhibition of MCAD and decreased octanoylcarnitine&malate stimulated respiration by approximately 60% (Osiki 2016 FASEB J). | |
| 3-Mercaptopropionic acid | MPA | 3-Mercaptopropionic acid (MPA) inhibits long chain acyl-CoA dehydrogenases (ACADs). |
| ADP | D | Adenosine diphosphate is a nucleotide. In OXPHOS core metabolism, ADP is a substrate of ANT and ATP synthase in the phosphorylation system. ADP is the discharged or low-energy counterpart of ATP. ADP can accept chemical energy by regaining a phosphate group to become ATP, in substrate-level phosphorylation (in anaerobic catabolism), at the expense of solar energy (in photosynthetic cells) or chemiosmotic energy (respiration in heterotrophic cells). ADP is added to mitochondrial preparations at kinetically saturating concentrations to induce the active state for evaluation of OXPHOS capacity. |
| AMPK | AMPK | AMP-activated protein kinase is a regulatory protein which acts as crucial cellular energy sensor by sensing AMP, ADP and/or Ca2+ levels in response to metabolic stresses or drug administration. |
| ASAPbio | Science only progresses as quickly and efficiently as it is shared. But even with all of the technological capabilities available today, the process of publishing scientific work is taking longer than ever. ASAPbio (Accelerating Science and Publication in biology) is a scientist-driven nonprofit working to address this problem by promoting innovation and transparency in life sciences communication. In 2015, ASAPbio founder Ron Vale published an analysis of the increasing time to first-author publication among graduate students at UCSF, and proposed a more widespread use of preprints in the life sciences as a potential solution. | |
| ATP | T | Adenosine triphosphate is a nucleotid and functions as the major carrier of chemical energy in the cells. As it transfers its energy to other molecules, it looses its terminal phosphate group and becomes adenosine diphosphate (ADP). |
| ATP synthase | CV | ATP synthase or F-ATPase (F1FO-ATPase; the use of Complex V is discouraged) catalyzes the endergonic phosphorylation of ADP to ATP in an over-all exergonic process that is driven by proton translocation along the protonmotive force. The ATP synthase can be inhibited by oligomycin. |
| ATPases | ATPases are enzymes that hydrolyse ATP, releasing ADP and inorganic phosphate. The contamination of isolated mitochondria with ATPases from other organelles and endogenous adenylates can lead to the production of ADP, which can stimulate respiration. This situation would lead to an overestimation of LEAK respiration measured in the absence of ADP, L(n) and subsequent inhibition of respiration by oligomycin, L(Omy). | |
| Abscissa | x | The abscissa is the horizontal axis x of a rectangular two-dimensional graph with the ordinate y as the vertical axis. Values X are placed horizontally from the origin. See Abscissal X/Y regression. |
| Absorbance | A | Also known as attenuation or extinction, absorbance (A) is a measure of the difference between the incident light intensity (I0) and the intensity of light emerging from a sample (I). It is defined as: A = log (I0/I) |
| Absorbance spectrum | When light enters a sample, the amount of light that it absorbs is dependent upon the wavelength of the incident light. The absorbance spectrum is the curve derived by plotting the measured absorbance against the wavelength of the light emerging from the sample over a given wavelength range. An absorbance spectrum may be characterised by peaks and troughs (absorbance maxima and minima) that can be used to identify, and sometimes quantify, different absorbing substances present in a sample. | |
| Absorption | Abs | When light enters a sample and emerges with an intensity (I), absorption (Abs) is the fraction of the light absorbed by the sample compared with the incident light intensity (I0): Abs = 1-I/I0. Absorption can also be expressed as Abs = 1-T, where T is the transmittance. |
| Absorption spectrum | An absorption spectrum is similar to an absorbance spectrum of a sample, but plotted as a function of absorption against wavelength. | |
| Abundance | In chemistry or physics, abundance or natural abundance refers to the amount of a chemical element isotope existing in nature. The abundance of an isotope on the Earth may vary depending on the place, but remains relatively constant in time (on a short-term scale). In a chemical reaction, the reactant is in abundance when the quantity of a substance is enough (or high) and constant during the reaction. Relative abundance represents the percentage of the total amount of all isotopes of the element. The relative abundance of each isotope in a sample can be identified using mass spectrometry. | |
| Acceleration | a, g [m·s-2] | Acceleration, a, is the change of velocity over time [m·s-2].
a = dv/dtThe symbol g is used for acceleration of free fall. The standard acceleration of free fall is defined as gn = 9.80665 [m·s-2]. |
| Acclimation | Acclimation is an immediate time scale adaption expressing phenotypic plasticity in response to changes of a single variable under controlled laboratory conditions. | |
| Acclimatization | Acclimatization is an immediate time scale adaption expressing phenotypic plasticity in response to changes of habitat conditions and life style where several variables may change simultaneously. | |
| Accuracy | The accuracy of a method is the degree of agreement between an individual test result generated by the method and the true value. | |
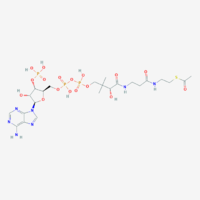
| Acetyl-CoA | Acetyl-CoA, C23H38N7O17P3S, is a central piece in metabolism involved in several biological processes, but its main role is to deliver the acetyl group into the TCA cycle for its oxidation. It can be synthesized in different pathways: (i) in glycolysis from pyruvate, by pyruvate dehydrogenase, which also forms NADH; (ii) from fatty acids β-oxidation, which releases one acetyl-CoA each round; (iii) in the catabolism of some amino acids such as leucine, lysine, phenylalanine, tyrosine and tryptophan.
In the mitochondrial matrix, acetyl-CoA is condensed with oxaloacetate to form citrate through the action of citrate synthase in the tricarboxylic acid cycle. Acetyl-CoA cannot cross the mitochondrial inner membrane but citrate can be transported out of the mitochondria. In the cytosol, citrate can be converted to acetyl-CoA and be used in the synthesis of fatty acid, cholesterol, ketone bodies, acetylcholine, and other processes. | |
| Aconitase | Aco | Aconitase is a TCA cycle enzyme that catalyzes the reversible isomerization of citrate to isocitrate. Also, an isoform is also present in the cytosol acting as a trans-regulatory factor that controls iron homeostasis at a post-transcriptional level. |
| Activity | a | The activity (relative activity) is a dimensionless quantity related to the concentration or partial pressure of a dissolved substance. The activity of a dissolved substance B equals the concentration, cB [mol·L-1], at high dilution divided by the unit concentration, c° = 1 mol·L-1:
aB = cB/c° This simple relationship applies frequently to substances at high dilutions <10 mmol·L-1 (<10 mol·m-3). In general, the concentration of a solute has to be corrected for the activity coefficient (concentration basis), γB, aB = γB·cB/c° At high dilution, γB = 1. In general, the relative activity is defined by the chemical potential, µB aB = exp[(µB-µ°)/RT] |
| Acyl-CoA dehydrogenase | ACAD | Acyl-CoA dehydrogenases ACADs are localized in the mitochondrial matrix. Several ACADs are distinguished: short-chain (SCAD), medium-chain (MCAD), and long-chain (LCAD). ACAD9 is expressed in human brain. ACADs catalyze the reaction
|
| Acyl-CoA oxidase | Acyl-CoA oxidase is considered as a rate-limiting step in peroxysomal β-oxidation, which carries out few β-oxidation cycles, thus shortening very-long-chain fatty acids (>C20). Electrons are directly transferred from FADH2 to O2 with the formation of H2O2. | |
| Acylcarnitine | AC | Acylcarnitines are esters derivative of carnitine and fatty acids, involved in the metabolism of fatty acids. Long-chain acylcarnitines such as palmitoylcarnitine must be transported in this form, conjugated to carnitine, into the mitochondria to deliver fatty acids for fatty acid oxidation and energy production. Medium-chain acylcarnitines such as octanoylcarnitine are also frequently used for high-resolution respirometry. |
| Adaptation | Adaptation is an evolutionary time scale expression of phenotypic plasticity in response to selective pressures prevailing under various habitat conditions. | |
| Add Graph/Delete bottom graph | Add: A new graph is added at the bottom of the screen. Select plots for display in the new graph, Ctrl+F6. Delete: Delete one of the graphs displayed in DatLab. | |
| Additive effect of convergent electron flow | Aα&β | Additivity Aα&β describes the principle of substrate control of mitochondrial respiration with convergent electron flow. The additive effect of convergent electron flow is a consequence of electron flow converging at the Q-junction from respiratory Complexes I and II (NS or CI&II e-input). Further additivity may be observed by convergent electron flow through glycerophosphate dehydrogenase and electron-transferring flavoprotein Complex. Convergent electron flow corresponds to the operation of the TCA cycle and mitochondrial substrate supply in vivo. Physiological substrate combinations supporting convergent NS e-input are required for reconstitution of intracellular TCA cycle function. Convergent electron flow simultaneously through Complexes I and II into the Q-junction supports higher OXPHOS capacity and ET capacity than separate electron flow through either CI or CII. The convergent NS effect may be completely or partially additive, suggesting that conventional bioenergetic protocols with mt-preparations have underestimated cellular OXPHOS-capacities, due to the gating effect through a single branch. Complete additivity is defined as the condition when the sum of separately measured respiratory capacities, N + S, is identical to the capacity measured in the state with combined substrates, NS (CI&II). This condition of complete additivity, NS=N+S, would be obtained if electron channeling through supercomplex CI, CIII and CIV does not interact with the pool of redox intermediates in the pathway from CII to CIII and CIV, and if the capacity of the phosphorylation system does not limit OXPHOS capacity (excess E-P capacity factor is zero). In most cases, however, additivity is incomplete, NS < N+S. |
| Adenine nucleotide translocase | ANT | The adenine nucleotide translocator, ANT, exchanges ADP for ATP in an electrogenic antiport across the inner mt-membrane. The ANT is inhibited by atractyloside, carboxyatractyloside and bongkrekik acid. The ANT is a component of the phosphorylation system. |
| Adenine nucleotides | AN | Adenine nucleotides, which are also sometimes referred to as adenosines or adenylates, are a group of organic molecules including AMP, ADP and ATP. These molecules present the major players of energy storage and transfer. |
| Adenylate kinase | ADK | Adenylate kinase, which is also called myokinase, is a phosphotransferase enzyme that is located in the mitochondrial intermembrane space and catalyzes the rephosphorylation of AMP to ADP in the reaction ATP + AMP ↔ ADP + ADP. |
| Advancement | dtrξ [MU] | In an isomorphic analysis, any form of flow is the advancement of a process per unit of time, expressed in a specific motive unit [MU∙s-1], e.g., ampere for electric flow or current, Iel = delξ/dt [A≡C∙s-1], watt for thermal or heat flow, Ith = dthξ/dt [W≡J∙s-1], and for chemical flow of reaction, Ir = drξ/dt, the unit is [mol∙s-1] (extent of reaction per time). The corresponding motive forces are the partial exergy (Gibbs energy) changes per advancement [J∙MU-1], expressed in volt for electric force, ΔelF = ∂G/∂elξ [V≡J∙C-1], dimensionless for thermal force, ΔthF = ∂G/∂thξ [J∙J-1], and for chemical force, ΔrF = ∂G/∂rξ, the unit is [J∙mol-1], which deserves a specific acronym [Jol] comparable to volt [V]. For chemical processes of reaction (spontaneous from high-potential substrates to low-potential products) and compartmental diffusion (spontaneous from a high-potential compartment to a low-potential compartment), the advancement is the amount of motive substance that has undergone a compartmental transformation [mol]. The concept was originally introduced by De Donder [1]. Central to the concept of advancement is the stoichiometric number, νi, associated with each motive component i (transformant [2]).
In a chemical reaction r the motive entity is the stoichiometric amount of reactant, drni, with stoichiometric number νi. The advancement of the chemical reaction, drξ [mol], is defined as, drξ = drni·νi-1 The flow of the chemical reaction, Ir [mol·s-1], is advancement per time, Ir = drξ·dt-1 This concept of advancement is extended to compartmental diffusion and the advancement of charged particles [3], and to any discontinuous transformation in compartmental systems [2], |
| Advancement per volume | dtrY [MU∙L-1] | Advancement per volume or volume-specific advancement, dtrY, is related to advancement of a transformation, dtrY = dtrξ∙V-1 [MU∙L-1]. Compare dtrY with the amount of substance j per volume, cj (concentration), related to amount, cj = nj∙V-1 [mol∙V-1]. Advancement per volume is particularly introduced for chemical reactions, drY, and has the dimension of concentration (amount per volume [mol∙L-1]). In an open system at steady-state, however, the concentration does not change as the reaction advances. Only in closed systems and isolated systems, specific advancement equals the change in concentration divided by the stoichiometric number, drY = dcj/νj (closed system) drY = drcj/νj (general) With a focus on internal transformations (i; specifically: chemical reactions, r), dcj is replaced by the partial change of concentration, drcj (a transformation variable or process variable). drcj contributes to the total change of concentration, dcj (a system variable or variable of state). In open systems at steady-state, drcj is compensated by external processes, decj = -drcj, exerting an effect on the total concentration change of substance j, dcj = drcj + decj = 0 (steady state) dcj = drcj + decj (general) |
| Advantage of preprints | The advantages of preprints, the excitement and concerns about the role that preprints can play in disseminating research findings in the life sciences are discussed by N Bhalla (2016). | |
| Aerobic | ox | The aerobic state of metabolism is defined by the presence of oxygen (air) and therefore the potential for oxidative reactions (ox) to proceed, particularly in oxidative phosphorylation (OXPHOS). Aerobic metabolism (with involvement of oxygen) is contrasted with anaerobic metabolism (without involvement of oxygen): Whereas anaerobic metabolism may proceed in the absence or presence of oxygen (anoxic or oxic conditions), aerobic metabolism is restricted to oxic conditions. Below the critical oxygen pressure, aerobic ATP production decreases. |
| Affinity of reaction | A [J·mol-1] | The concept of affinity and hence chemical force is deeply rooted in the notion of attraction (and repulsion) of alchemy, which was the foundation of chemistry originally, but diverted away from laboratory experiments towards occult secret societies [1].** Newton's extensive experimental alchemical work and his substantial written track record on alchemy (which he did not publish) is seen today as a key inspiration for his development of the concept of the gravitational force [2-4]. This marks a transition of the meaning of affinity, from the descriptive 'adjacent' (proximity) to the causative 'attractive' (force) [5]. Correspondingly, Lavoisier (1790) equates affinity and force [6]: “... the degree of force or affinity with which the acid adheres to the base” [5]. By discussing the influence of electricity and gravity on chemical affinity, Liebig (1844) considers affinity as a force [7]. This leads to Guldberg and Waage's mass action ratio ('Studies concerning affinity', 1864; see [5]), the free energy and chemical affinity of Helmholtz (1882 [8]), and chemical thermodynamics of irreversible processes [9], where flux-force relations are center stage [10].
According to the IUPAC definition, the affinity of reaction, A [J·mol-1], equals the negative molar Gibbs energy of reaction [11], which is the negative Gibbs force of reaction (derivative of Gibbs energy per advancement of reaction [12]): -A = ΔrF = ∂G/∂rξThe historical account of affinity is summarized by concluding, that today affinity of reaction should be considered as an isomorphic motive force and be generalized as such. This will help to (1) avoid confusing reversals of sign conventions (repulsion = negative attraction; pull = negative push), (2) unify symbols across classical and nonequilibrium thermodynamics [12,13], and thus (3) facilitate interdisciplinary communication by freeing ourselves from the alchemical, arcane scientific nomenclature. |
| Air calibration | R1 | Air calibration of an oxygen sensor (polarographic oxygen sensor) is performed routinely on any day before starting a respirometric experiment. The volume fraction of oxygen in dry air is constant. An aqueous solution in equilibrium with air has the same partial pressure as that in water vapour saturated air. The water vapour is a function of temperature only. The partial oxygen pressure in aqueous solution in equilibrium with air is, therefore, a function of total barometric pressure and temperature. Bubbling an aqueous solution with air generates deviations from barometric pressure within small gas bubbles and is, therefore, not recommended. To equilibrate an aqueous solution ata known partial pressure of oxygen [kPa], the aqueous solution is stirred rigorously in a chamber enclosing air at constant temperature. The concentration of oxygen, cO2 [µM], is obtained at any partial pressure by multiplying the partial pressure by the oxygen solubility, SO2 [µM/kPa]. SO2 is a function of temperature and composition of the salt solution, and is thus a function of the experimental medium. The solubility factor of the medium, FM, expresses the oxygen solubility relative to pure water at any experimental temperature. FM is 0.89 in serum (37 °C) and 0.92 in MiR06 or MiR05 (30 °C and 37 °C). |
| Allegations of research misconduct | Allegations of research misconduct are handled with care. Publishers and editors shall take reasonable steps to identify and prevent the publication of papers where research misconduct has occurred, including plagiarism, citation manipulation, and data falsification/fabrication, among others. In no case shall a journal or its editors encourage such misconduct, or knowingly allow such misconduct to take place. In the event that a journal's publisher or editors are made aware of any allegation of research misconduct relating to a published article in their journal, the publisher or editor shall follow COPE's guidelines (or equivalent) in dealing with allegations. | |
| Alternative oxidase | AOX | Alternative quinol oxidases AOX are membrane-bound enzymes capable of supporting cyanide- and antimycin A-resistant mitochondrial respiration. AOX catalyzes the oxidation of ubiquinol and the reduction of oxygen to water in a four-electron process. As this bypasses several proton-translocating steps, induction of this alternative pathway is associated with a reduction of ATP production per oxygen consumed. AOX is found in most plants (including microalgae), many fungi and protists, but is not expressed in animals. AOX is inhibited by salicylhydroxamic acid (SHAM). Expression and activity of the enzyme are modified by environmental conditions such as temperature, oxidative stress, nutrient availability, and pathogens such as viruses. |
| Ambiguity crisis |
The ambiguity crisis is a contemporary crisis comparable to the credibility or reproducibility crisis in the biomedical sciences. The term 'crisis' is rooted etymologically in the Greek word krinein: meaning to 'separate, decide, judge'. In this sense, science and communication in general are a continuous crisis at the edge of separating clarity or certainty from confusing double meaning, or obscure 'alchemical' gibberish, or even fake-news. Reproducibility relates to the condition of repeating and confirming calculations or experiments presented in a published resource. While ambiguity is linked to relevant issues of reproducibility, it extends to the communications space of terminological and graphical representations of concepts. Type 1 ambiguities are the inevitable consequence of conceptual evolution, in the process of which ambiguities are replaced by experimentally and theoretically supported paradigm shifts to clear-cut theorems. In contrast, type 2 ambiguities are traced in publications that reflect merely a disregard and ignorance of established concepts without an attempt to justify the inherent deviations from high-quality science. There are many shades of grey between these types of ambiguity. | |
| Ammonia solution concentrated | NH3 | Concentrated ammonia solution (25 % - 30 % ammonium hydroxide solution, ammonia) is used for the service of the polarographic oxygen sensor OroboPOS. After opening the commercial solution, the concentration of ammonia may decline during storage and may render the ammonia stock ineffective for sensor service. Source: A commercially available solution from a drugstore is sufficient for this cleaning purpose |
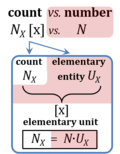
| Amount of substance | n [mol] | The amount of substance n is a base physical quantity, and the corresponding SI unit is the mole [mol]. Amount of substance (sometimes abbreviated as 'amount' or 'chemical amount') is proportional to the number NX of specified elementary entities X, and the universal proportionality constant is the reciprocal value of the Avogadro constant (SI),
nX = NX·NA-1 nX contained in a system can change due to internal and external transformations, dnX = dinX + denX In the absence of nuclear reactions, the amount of any atom is conserved, e.g., for carbon dinC = 0. This is different for chemical substances or ionic species which are produced or consumed during the advancement of a reaction r, A change in the amount of Xi, dni, in an open system is due to both the internal formation in chemical transformations, drni, and the external transfer, deni, across the system boundaries. dni is positive if Xi is formed as a product of the reaction within the system. deni is negative if Xi flows out of the system and appears as a product in the surroundings (Cohen 2008 IUPAC Green Book). |
| Amp calibration - DatLab | F5 | Amp calibration indicates the calibration of the amperometric O2k-channel. |
| Ampere | A | The ampere, symbol A, is the SI unit of electric current. It is defined by taking the fixed numerical value of the elementary charge e to be 1.602 176 634 × 10−19 when expressed in the unit C, which is equal to A s, where the second is defined in terms of ΔνCs. |
| Amperometric,Amp | F7 | After selection of the Amperometric, Amp channel in the O2k configuration, an Amperometric, Amp tab will appear in the O2k control [F7] window. Set the desired light intensity (0-1600) in the field ´Fluo intensity´ and the desired amplification of the signal (1-1000) in the field ´Gain for Fluo sensor´in the Amperometric, Amp window followed by a left-click Send to O2k. Switching off the illumination before each fluorometric measurement is routinely required. |
| Amplex UltraRed | AmR | Amplex® UltraRed (AmR) is used as an extrinsic fluorophore for measurement of hydrogen peroxide production (ROS) by cells or mitochondrial preparations. The reaction of H2O2 and AmR is catalyzed by horseradish peroxidase to produce the red fluorescent compound resorufin (excitation wavelength 563 nm, emission 587 nm; the fluorescent product according to the supplier is called UltroxRed in the case of Amplex® UltraRed which has a similar structure to resorufin). The change of emitted fluorescence intensity is directly proportional to the concentration of H2O2 added, whereby the H2O2 is consumed. |
| Amplitude | The amplitude of the absorbance spectrum can be described in terms of the absorbance differences between the characteristic peaks (absorbance maxima) and troughs (absorbance minima) (see absorbance spectrum) for substances present in the sample. | |
| Amytal | Amy | Amytal sodium salt (synonym: amobarbital; 5-Ethyl-5-isoamylbarbituric acid) is a barbiturate drug and an inhibitor of Complex I. |
| Anaerobic | Anaerobic metabolism takes place without the use of molecular oxygen, in contrast to aerobic metabolism. The capacity for energy assimilation and growth under anoxic conditions is the ultimate criterion for facultative anaerobiosis. Anaerobic metabolism may proceed not only under anoxic conditions or states, but also under hyperoxic and normoxic conditions (aerobic glycolysis), and under hypoxic and microxic conditions below the limiting oxygen pressure. | |
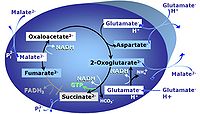
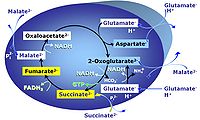
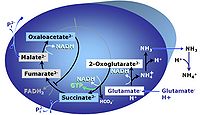
| Anaplerosis | Anaplerosis is the process of formation of intermediates of the tricarboxylic acid cycle. Malic enzyme (mtME), phosphoenolpyruvate carboxykinase (PEPCK), propionyl-CoA carboxylase, pyruvate carboxylase and proline dehydrogenase play important roles in anaplerosis. | |
| Anaplerotic pathway control state | a | Anaplerotic pathway control states are fuelled by single substrates which are transported into the mitochondrial matrix and increase the pool of intermediates of the tricarboxylic acid cycle. Malic enzyme (mtME), phosphoenopyruvate carboxykinase (PEPCK), propionyl-CoA carboxylase, and pyruvate carboxylase play important roles in anaplerosis. The glutamate-anaplerotic pathway control state and malate-anaplerotic pathway control state are the most important anaplerotic substrate control states (aN). |
| Anoxia | anox | Ideally the terms anoxia and anoxic (anox, without oxygen) should be restricted to conditions where molecular oxygen is strictly absent. Practically, effective anoxia is obtained when a further decrease of experimental oxygen levels does not elicit any physiological or biochemical response. The practical definition, therefore, depends on (i) the techiques applied for oxygen removal and minimizing oxygen diffusion into the experimental system, (ii) the sensitivity and limit of detection of analytical methods of measuring oxygen (O2 concentration in the nM range), and (iii) the types of diagnostic tests applied to evaluate effects of trace amounts of oxygen on physiological and biochemical processes. The difficulties involved in defining an absolute limit between anoxic and microxic conditions are best illustrated by a logarithmic scale of oxygen pressure or oxygen concentration. In the anoxic state (State 5), any aerobic type of metabolism cannot take place, whereas anaerobic metabolism may proceed under oxic or anoxic conditions. |
| Antimycin A | Ama | Antimycin A is an inhibitor of Complex III (CIII). It binds to the Qi site of CIII and inhibits the transfer of electrons from heme bH to oxidized Q (Qi site inhibitor). High concentrations of antimycin A also inhibit acyl-CoA oxidase and D-amino acid oxidase. |
| Ap5A | Ap5A | P1,P5-Di(adenosine-5')pentaphosphate (Ap5A) is an inhibitor of adenylate kinase (ADK), the enzyme which rephosphorylates AMP to ADP, consuming ATP (ATP + AMP ↔ 2 ADP). |
| Aqua destillata | a.d. | Aqua destillata (a.d.) is the Latin name for distilled water, H2O. When a.d. is used in various solution protocols, it may indicate that water with the highest possible quality or lowest possible level of impurities should be used, as may be reached not only with distilled water but also with high-purity deionised water. |
| ArXiv preprint server | arXiv | arXiv is a classic preprint server initiated in 1991 by Paul Ginsparg. {Quote} arXiv.org is a highly-automated electronic archive and distribution server for research articles. Covered areas include physics, mathematics, computer science, nonlinear sciences, quantitative biology, quantitative finance, statistics, electrical engineering and systems science, and economics. arXiv is maintained and operated by Cornell University with guidance from the arXiv Scientific Advisory Board and the arXiv Member Advisory Board, and with the help of numerous subject moderators. {end of Quote}. arXiv rejects abstracts that are submitted without accompanying paper. |
| Artemisinin | Artemisinin and various derivatives are potent anti-malaria drugs which have additionally anti-tumorigenic effects, particularly when targeted at mitochondria. The anti-malaria effect is associated with artemisinin's action on heme. Mitochondria are involved in the synthesis of heme, and may play additional roles in the anti-tumorigenic effect of artemisinin. | |
| Ascorbate | As | In respiratory assays for cytochrome c oxidase activity (Complex IV, CIV), ascorbate is added as regenerating system to maintain TMPD in a reduced state. It has to be titrated into the respiration medium prior to the addition of TMPD, otherwise the autoxidation reaction velocity is permanently elevated. |
| Asia Society for Mitochondrial Research and Medicine | ASMRM | The Asia Society for Mitochondrial Research and Medicine (ASMRM) was founded in 2003 to share the latest knowledge on mitochondrial research. |
| Aspirin | Aspirin is a widely applied drug that requires dosage adjusted to individual body mass. It is a non-selective COX inhibitor and exerts an effect on long-chain fatty acid transport into mitochondria. | |
| Assay | An experimental assay is a method to obtain a measurement with a defined instrument on a sample or subsample. Multiple assay types may be applied on the same sample or subsample, if the measurement does not destroy it. For instance, the wet weight of a permeabilized muscle fibre preparation can be determined based on a specific laboratory protocol (gravimetric assay), maintaining the functional integrity of the sample, which then can be used in a respirometric assay, followed by a spectrophotometric assay for measurement of protein content. The experimental design determines which types of assays have to be applied for a complete experiment. Destructive assays, such as determination of protein content or dry weight, can be applied on a sample only after performing a respirometric assay, or on a separate subsample. The experimental variability is typically dominated by the assay with the lowest resolution or signal to noise ratio. The signal to noise ratio may be increased by increasing the number, n, of repetitions of measurements on subsamples. Evaluation of procedural variation ('experimental noise') due to instrumental resolution and handling requires subsampling from homogenous samples. | |
| Atractyloside | Atr | Atractyloside is an inhibitor of the adenine nucleotide translocator (ANT). It is an extremely toxic glycoside that inhibits oxidative phosphorylation by blocking the transfer of adenosine nucleotides through the mitochondrial membrane. |
| Attached cells | Many cell types are grown in culture as attached cells, such as endothelial or neuronal cells in a monolayer. | |
| Attribute | Attribute in general is a characteristic or property. In databases an attribute describes a column in a table. Rows then represent the according attribute values. | |
| Auranofin | AF | Auranofin (AF) is a gold complex which inhibites thioredoxin reductase (TrxR). |
| Automatic pan - DatLab | Automatic pan (only for real-time data recording) toggles automatic panning on/off by clicking in the O2k status line. If it is on (green), the time range is maintained while the time axis always shows the currently recorded data, i.e. the value of the offset (minimum value) increases as experimental time proceeds. If it is off (yellow), the time axis is static. This allows for manually panning backwards to observe previous sections of the experiment at a given time range. In this mode, the actual experimental time may be off-scale. Toggle between "Pan auto" and "Pan off" by a left-click on the text. It does not influence continuous data recording. It is recommended to maintain automatic panning on during the experiment, except for specifically viewing earlier sections of the experiment. | |
| Autoscale | Autoscale zooms in or out of the selected period with Autoscale time axis, Autoscale Y1 (Y2) axes and Automatic pan. | |
| Autoscale Y1 (Y2) axes | Autoscale Y1 (Y2) axes: Autoscaling the measured values (full data range) on the Y1 (Y2) axis in the selected plot. | |
| Autoscale time axis | Autoscale time axis gives an overview of the entire experimental period. | |
| Autoxidation | This definition is insufficient and needs elaboration. Autoxidation is a slow process implying oxidation of carbohydrates through oxygen in open air, leading to a primary formation of peroxides and hydroperoxides. UV radiation can speed up this process. | |
| Averaging | In order to improve the signal-to-noise ratio a number of sequential spectra may be averaged over time. The number of spectra to be averaged can be set prior to carrying out the measurements, or afterwards during data analysis. | |
| Avogadro constant | NA [x·mol-1] | {Quote} The Avogadro constant NA is a proportionality constant between the quantity amount of substance (with unit mole) and the quantity for counting entities ... One mole contains exactly 6.022 140 76 × 1023 elementary entities. This number is the fixed numerical value of the Avogadro constant, NA, when expressed in the unit mol−1 and is called the Avogadro number {End of Quote: Bureau International des Poids et Mesures 2019 The International System of Units (SI)}. Thus the Avogadro constant NA has the SI unit 'per mole' [mol-1], but more strictly the unit for counting entities per amount is 'units per mole' [x·mol-1] (compare elementary charge). Therefore, NA is 'count per amount' with units 'counting units per mole'. The Avogadro constant times elementary charge is the Faraday constant. |
| Azide | Azd | Sodium azide is an inhibitor of Complex IV/cytochrome c oxidase (CIV, COX, CcO). |
| BAM15 | BAM15 | 2-fluorophenyl){6-[(2-fluorophenyl)amino](1,2,5-oxadiazolo[3,4-e]pyrazin-5-yl)}amine (BAM15) is a protonophore or uncoupler of oxidative phosphorylation detected in a screen for uncoupling agents exerting less toxicity than commonly used uncouplers and first described by Kennwood et al. 2013. In their comparison of BAM15 with FCCP it was shown to increase oxygen flux to a similar extent as the classical uncoupler, to display a much broader range of concentrations inducing maximum respiration, to stimulate no formation of H2O2, to leave cellular membrane potential unaffected, and to ultimately exert less cytotoxicity. |
| BME cutoff points | BME cutoff | Obesity is defined as a disease associated with an excess of body fat with respect to a healthy reference condition. Cutoff points for body mass excess, BME cutoff points, define the critical values for underweight (-0.1 and -0.2), overweight (0.2), and various degrees of obesity (0.4, 0.6, 0.8, and above). BME cutoffs are calibrated by crossover-points of BME with established BMI cutoffs. |
| Background state | Y | The background state Y (background rate YX) is the non-activated or inhibited respiratory state at background rate, which is low in relation to the higher rate ZX in the reference state Z. The transition from the background state to the reference state is a step change. A metabolic control variable X (substrate, activator) is added to the background state to stimulate flux to the level of the reference state. Alternatively, the metabolic control variable X is an inhibitor, which is present in the background state Y, but absent in the reference state Z. The background state is the baseline of a single step in the definition of the flux control efficiency. In a sequence of step changes, the common baseline state is the state of lowest flux in relation to all steps, which can be used as a baseline correction. |
| Balance | In transmission spectrophotometry blank cuvettes are used to record the incident light intensity (I0) prior to absorbance measurements. (See white balance for reflectance spectrophotometry, remittance spectrophotometry). | |
| Bandwidth | Bandwidth is measured in nanometers in terms of the full width half maximum of a peak. This is the portion of the peak that is greater than half of the maximum intensity of that peak. | |
| Barometric pressure | pb [Pa] | Barometric pressure, pb, is an important variable measured for calibration of oxygen sensors in solutions equilibrated with air. The atm-standard pressure (1 atm = 760 mmHg = 101.325 kPa) has been replaced by the SI standard pressure of 100 kPa. The partial pressure of oxygen, pO2, in air is a function of barometric pressure, which changes with altitude and locally with weather conditions. The partial oxygen pressure declines by 12 % to 14 % per 1,000 m up to 6,000 m altitude, and by 15 % to 17 % per 1,000 m between 6,000 and 9,000 m altitude. The O2k-Barometric Pressure Transducer is built into the Oroboros O2k as a basis for accurate air calibrations in high-resolution respirometry. For highest-level accuracy of calculation of oxygen pressure, it is recommended to compare at regular intervals the barometric pressure recording provided by the O2k with a calibrated barometric pressure recording at an identical time point and identical altitude. The concept of gas pressure or barometric pressure can be related to the generalized concept of isomorphic pressure. |
| Barth Syndome | BTHS | Barth Syndome (BTHS) is an X-linked genetic condition that is caused by a mutation in the tafazzin gene (taz). This mutation causes cardiolipin abnormalities, cardiomyopathy, neutropenia, muscle weakness, growth delay, and exercise intolerance. Contributed by Sparagna GC 2016-04-24 |
| Basal respiration | BMR | Basal respiration or basal metabolic rate (BMR) is the minimal rate of metabolism required to support basic body functions, essential for maintenance only. BMR (in humans) is measured at rest 12 to 14 hours after eating in a physically and mentally relaxed state at thermally neutral room temperature. Maintenance energy requirements include mainly the metabolic costs of protein turnover and ion homeostasis. In many aerobic organisms, and particularly well studied in mammals, BMR is fully aerobic, i.e. direct calorimetry (measurement of heat dissipation) and indirect calorimetry (measurement of oxygen consumption multiplied by the oxycaloric equivalent) agree within errors of measurement (Blaxter KL 1962. The energy metabolism of ruminants. Hutchinson, London: 332 pp [1]). In many cultured mammalian cells, aerobic glycolysis contributes to total ATP turnover (Gnaiger and Kemp 1990 [2]), and under these conditions, 'respiration' is not equivalent to 'metabolic rate'. Basal respiration in humans and skeletal muscle mitochondrial function (oxygen kinetics) are correlated (Larsen et al 2011 [3]). » MiPNet article |
| Base quantities and count | Template:Base quantities and count | |
| Baseline state | The baseline state in a sequence of step changes is the state of lowest flux in relation to all steps, which can be used as a baseline correction. Correction for residual oxygen consumption, ROX, is an example where ROX is the baseline state. In a single step, the baseline state is equivalent to the background state. | |
| Beer-Lambert law | B-L law | This law states that the transmittance (T) of light though a sample is given by: T = e-εbc, where ε is the molar extinction coefficient, b is the pathlength of the light through the cuvette (in mm) and c is the concentration of the pigment in the sample (in mM). Transforming this equation, it can be seen that the absorbance of light (A) is simply given by A = εbc. |
| Beryllium sulfate | BeS | Beryllium sulfate is used in combination with sodium fluoride to form beryllium trifluoride (BeF3−), to inhibit the ATP synthase if it is exposed by disruption of the mitochondrial membranes. |
| Bias | The bias is defined as the difference between the mean of the measurements and the reference value. In general, the measuring instrument calibration procedures should focus on establishing and correcting it. | |
| BioRxiv preprint server for biology | bioRxiv | bioRxiv (pronounced "bio-archive") is a free online archive and distribution service for unpublished preprints in the life sciences. It was launched in 2013 by Cold Spring Harbor Laboratory Press in New York, and is operated by Cold Spring Harbor Laboratory, a not-for-profit research and educational institution. By posting preprints on bioRxiv, authors are able to make their findings immediately available to the scientific community and receive feedback on draft manuscripts before they are submitted to journals. bioRxiv is intended for rapid sharing of new research. Some review articles contain new data/analyses and may therefore be deemed appropriate. Reviews that solely summarize existing knowledge are not appropriate and neither are term papers, book excerpts, and undergraduate dissertations. |
| Bioblast | ||
| Bioblast alert 2020 | ||
| Bioblast alert 2021 | ||
| Bioblast alert 2022 | ||
| Bioblast alert 2023 | ||
| Bioblast alert 2024 | ||
| Bioblast track | ||
| Bioblast:About | ||
| Bioblasts | BB | Richard Altmann (1894) defined the 'elementary organisms' as Bioblasts. He observed granula in cells stained with osmium and viewed ‘the protoplasm as a colony of bioblasts’. "Microorganisms and granula are at an equivalent level and represent elementary organisms, which are found wherever living forces are acting, thus we want to describe them by the common term bioblasts. In the bioblast, that morphological unit of living matter appears to be found." Altmann 1894; p. 141. Altmann is thus considered as the discoverer of mitochondria (the granula), which constitute together with the microorganisms the bioblasts (the elementary organisms). Bioblasts are the aliens with permanent residence in our cells (Gnaiger 2010). |
| Biochemical coupling efficiency | jE-L | |
| Biochemical threshold effect | Due to threshold effects, even a large defect diminishing the velocity of an individual enzyme results in only minor changes of pathway flux. | |
| Biological contamination | Biological contamination may be caused by microbial growth in the O2k-Chamber or in the experimental medium. | |
| Biological reference interval | Biological reference interval or reference interval is the central 95 % interval of the distribution of reference values. | |
| Biopsy preservation solution | BIOPS | Biopsy preservation solution, for preservation of tissue samples, preparation of muscle fibres, and permeabilization with saponin. |
| Blank | In fluorometry and transmission spectrophotometry blank cuvettes (with no samples in them) are used to carry out the balance. | |
| Blebbistatin | Bleb | Blebbistatin is a widely used muscle and non-muscle myosin II-specific inhibitor that block contractile activity. Blebbistatin shows selectivity and high affinity for multiple class II myosins. Blebbistatin is commonly employed in respirometric experiments with permeabilized muscle fibers (pfi). Permeabilized muscle fibers are sensitive to low oxygen supply due to diffusion restrictions that limit mitochondrial respiration at the core of the fiber bundle. Therefore, hyperoxic conditions are required to counteract this limitation. Further studies have shown that the addition of blebbistatin in the respiration medium prevents fiber contraction, reduces the oxygen sensitivity and allows the study of ADP kinetics in pfi at normoxic oxygen levels. However, other studies described that the presence of blebbistatin does not prevent the oxygen dependence in pfi. Moreover, several limitations of blebbistatin i.e. low solubility in water, cytotoxicity and phototoxicity have been described. |
| Block temperature | The block temperature of the Oroboros O2k is the continuously measured temperature of the copper block, housing the two glass chambers of the O2k. The block temperature is recorded by DatLab as one of the O2k system channels. | |
| Blood cell preparation | bcp | Blood cell preparation (bcp) is one of the key steps in diagnostic protocols. |
| Blood plasma | Plasma | Blood plasma is the non-cellular component of the blood. Plasma lacks cellular components of the blood, red blood cells, white blood cells, and platelets. However, there are many proteins in plasma, i.e. fibrinogen, albumin and globulin. Both blood plasma and platelet-rich plasma maintain clotting activity after whole blood separation. |
| Blood serum | Serum | Blood serum is a purified plasma in which the coagulant components were removed from the blood plasma. It contains other substances, i.e. antibodies, antigens and hormones. Serum can be obtained by collecting the liquid phase after blood or plasma coagulation. |
| Body fat excess | BFE | In the healthy reference population (HRP), there is zero body fat excess, BFE, and the fraction of excess body fat in the HRP is expressed - by definition - relative to the reference body mass, M°, at any given height. Importantly, body fat excess, BFE, and body mass excess, BME, are linearly related, which is not the case for the body mass index, BMI. |
| Body mass | m [kg]; M [kg·x-1] | The body mass M is the mass (kilogram [kg]) of an individual (object) [x] and is expressed in units [kg/x]. Whereas the body weight changes as a function of gravitational force (you are weightless at zero gravity; your floating weight in water is different from your weight in air), your mass is independent of gravitational force, and it is the same in air and water. |
| Body mass excess | BME | The body mass excess, BME, is an index of obesity and as such BME is a lifestyle metric. The BME is a measure of the extent to which your actual body mass, M [kg/x], deviates from M° [kg/x], which is the reference body mass [kg] per individual [x] without excess body fat in the healthy reference population, HRP. A balanced BME is BME° = 0.0 with a band width of -0.1 towards underweight and +0.2 towards overweight. The BME is linearly related to the body fat excess. |
| Body mass index | BMI | The body mass index, BMI, is the ratio of body mass to height squared (BMI=M·H-2), recommended by the WHO as a general indicator of underweight (BMI<18.5 kg·m-2), overweight (BMI>25 kg·m-2) and obesity (BMI>30 kg·m-2). Keys et al (1972; see 2014) emphasized that 'the prime criterion must be the relative independence of the index from height'. It is exactly the dependence of the BMI on height - from children to adults, women to men, Caucasians to Asians -, which requires adjustments of BMI-cutoff points. This deficiency is resolved by the body mass excess relative to the healthy reference population. |
| Boltzmann constant | k [J·x-1·K-1] | The Boltzmann constant k has the SI unit [J·K-1] (IUPAC), but more strictly the units for energy per particles per temperature is [J·x-1·K-1].
k = f·e-1, the electrochemical constant f times the elementary charge e. k = R·NA-1, the gas constant R divided by the Avogadro constant NA. |
| Bongkrekik acid | Bka | Bongkrekik acid is a selective and potent inhibitor of the adenine nucleotide translocator (ANT). Bka binds to the matrix (negative) site of ANT, opposite of carboxyatractyloside. |
| Bosy-Westphal 2009 Br J Nutr | ||
| Bound energy | B [J] | The bound energy change in a closed system is that part of the total energy change that is always bound to an exchange of heat,
dB = dU - dA [Eq. 1] ∆B = ∆H - ∆G [Eq. 2] The free energy change (Helmoltz or Gibbs; dA or dG) is the total energy change (total inner energy or enthalpy, dU or dH) of a system minus the bound energy change. Therefore, if a process occurs at equilibrium, when dG = 0 (at constant gas pressure), then dH = dB, and at deW = 0 (dH = deQ + deW; see energy) we obtain the definition of the bound energy as the heat change taking place in an equilibrium process (eq), dB = T∙dS = deQeq [Eq. 3] |
| Bovine serum albumin | BSA | Bovine serum albumin is a membrane stabilizer, oxygen radical scavenger, and binds Ca2+ and free fatty acids, hence the rather expensive essentially free fatty acid free BSA is required in mitochondrial isolation and respiration media. Sigma A 6003 fraction V. |
| Buffer Z | Buffer Z | Mitochondrial respiration medium, Buffer Z, described by Perry 2011 Biochem J For composition and comparison see: Mitochondrial respiration media: comparison |
| CDGSH iron-sulfur domain proteins | CISD proteins | The CDGSH iron-sulfur domain (CISDs) family of proteins uniquely ligate labile 2Fe-2S clusters with a 3Cys-1His motif. CISD1 and CISD3 have been demonstrated to localize to the outer mitochondrial membrane and mitochondrial matrix respectively, however their relationship to mitochondrial physiology remains ill-defined [1]. The best characterized member of the CISD family, CISD1, has been demonstrated to be involved in respiratory capacity, iron homeostasis, and ROS regulation |
| CE | CE | CE marking is a mandatory conformity marking for certain products sold within the European Economic Area (EEA). |
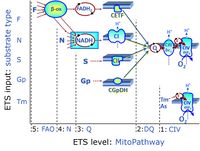
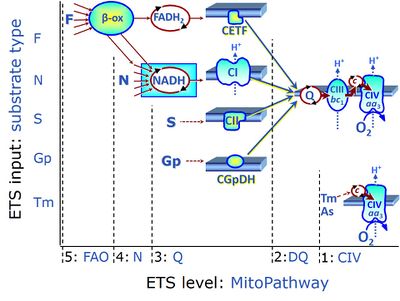
| CHNO-fuel substrate | CHNO | CHNO-fuel substrates are reduced carbon-hydrogen-nitrogen-oxygen substrates which are oxidized in the exergonic process of cell respiration. Mitochondrial pathways are stimulated by CHNO-fuel substrates feeding electrons into the ETS at different levels of integration and in the presence or absence of inhibitors acting on specific enzymes which are gate-keepers and control various pathway segments. |
| CI control ratio | N/NS; CI/CI&II | See N/NS pathway control ratio |
| CII control ratio | S/NS; CII/CI&II | See S/NS pathway control ratio |
| COPE core practices for research | COPE core practices for research are applicable to all involved in publishing scholarly literature. | |
| Calcium | Ca | Ca2+ is a major signaling molecule in both prokaryotes and eukaryotes. Its cytoplasmic concentration is tightly regulated by transporters in the plasma membrane and in the membranes of various organelles. For this purpose, it is either extruded from the cell through exchangers and pumps or stored in organelles such as the endoplasmic reticulum and the mitochondria. Changes in the concentration of the cation regulate numerous enzymes including many involved in ATP utilizing and in ATP generating pathways and thus ultimately control metabolic activity of mitochondria and of the entire cell. Measuring changes in Ca2+ levels is thus of considerable interest in the context of high-resolution respirometry. |
| Calcium Green | CaG | Calcium GreenTM (CaG) denotes a family of extrinsic fluorophores applied for measurement of Ca2+ concentration with mitochondrial preparations. This dye fluoresces when bound to Ca2+. When measuring mitochondrial calcium uptake it is possible to observe the increase of the CaG signal upon calcium titration, followed by the decrease of CaG signal due to the uptake. |
| Calcium retention capacity | CaRC | Calcium retention capacity (CaRC) is a measure of the capability of mitochondria to retain calcium (Ca2+), primarily in the form of calcium phosphates, in the mitochondrial matrix. By storing calcium in the form of osmotically inactive precipitates the mitochondria contribute to the buffering of cytosolic free Ca2+ levels and thereby to the regulation of calcium-dependent cellular processes. Alterations of CaRC are important in stress phenomena associated with energy limitation and have been linked to neurodegenerative diseases (Starkov 2013 FEBS J). Experimentally, CaRC has been indirectly assessed by determination of respiratory rates of isolated mitochondria which were exposed to continuously increasing doses of Ca2+ by use of the Titration-Injection microPump TIP2k. The upper limit of CaRC was observed as a sudden decrease of respiration presumed to reflect opening of the permeability transition pore (Hansson 2010 J Biol Chem). |
| Calorespirometric ratio | CR ratio [kJ/mol] | The calorimetric/respirometric or calorespirometric ratio (CR ratio) is the ratio of calorimetrically and respirometrically measured heat and oxygen flux, determinded by calorespirometry. The experimental CR ratio is compared with the theoretically derived oxycaloric equivalent, and agreement in the range of -450 to -480 kJ/mol O2 indicates a balanced aerobic energy budget (Gnaiger and Staudigl 1987). In the transition from aerobic to anaerobic metabolism, there is a limiting pO2, plim, below which CR ratios become more exothermic since anaerobic energy flux is switched on. |
| Calorespirometry | CR | Calorespirometry is the method of measuring simultaneously metabolic heat flux (calorimetry) and oxygen flux (respirometry). The calorespirometric ratio (CR ratio; heat/oxygen flux ratio) is thus experimentally determined and can be compared with the theoretical oxycaloric equivalent, as a test of the aerobic energy balance. |
| Candela | cd | The candela, symbol cd, is the SI unit of luminous intensity in a given direction. It is defined by taking the fixed numerical value of the luminous efficacy of monochromatic radiation of frequency 540 × 1012 Hz, Kcd, to be 683 when expressed in the unit lm W−1. |
| Canonical ensemble | A canonical ensemble is the group of compartments enclosed in an isolated system H, with a smaller compartment A1 in thermal equilibrium with a larger compartment A2 which is the heat reservoir at temperature T. When A1 is large in the canonical sense, if its state can be described in terms of macroscopic thermodynamic quantities of V, T, and p merging with the state described as a probability distribution. | |
| Carbohydrate | Carbohydrates, also known as saccharides, are molecules composed of carbon, hydrogen and oxygen. These molecules can be divided by size and complexity into monosaccharides, disaccharides, oligosaccharides, and polysaccharides. Glucose is a monosaccharide considered the primary source of energy in cells and a metabolic intermediate. This carbohydrate undergoes glycolysis, with the generation of pyruvate, that can enter the TCA cycle. Carbohydrates such as glucose and fructose may also be involved in the Crabtree effect. | |
| Carbonyl cyanide m-chlorophenyl hydrazone | CCCP | Carbonyl cyanide m-chlorophenyl hydrazone, CCCP (U; C9H5ClN4; FW = 204.62) is a protonophore (H+ ionophore) and is used as a potent chemical uncoupler of oxidative phosphorylation. Like all uncouplers, CCCP concentrations must be titrated carefully to evaluated the optimum concentration for maximum stimulation of mitochondrial respiration, particularly to avoid inhibition of respiration at higher CCCP concentrations. |
| Carboxy SNARF 1 | SNARF | Carboxy SNARF® 1 is a cell-impermeant pH indicator dye. The pKa of ~7.5 makes it useful for measuring pH in the range of pH 7 to pH 8. The emission shifts from yellow-orange at low pH to deep red fluorescence at high pH. Ratiometric fluorometry, therefore, is applied at two emission wavelengths,such as 580 nm and 640 nm. Relative molecular mass: Mr = 453.45 |
| Carboxyatractyloside | CAT | Carboxyatractyloside CAT is a highly selective and potent inhibitor of the adenine nucleotide translocator (ANT). CAT stabilizes the nucleoside binding site of ANT on the cytoplasmic (positive) side of the inner membrane and blocks the exchange of matrix ATP and cytoplasmic ADP. It causes stabilization of the c conformation of ANT leading to permeability transition pore (PTP) opening, loss of mitochondrial membrane potential, and apoptosis. |
| Cardiolipin | CL | Cardiolipin, CL, is a double phospholipid (having 4 fatty acyl chains) in the mitochondrial inner membrane (mtIM) which plays an important role in mitochondrial bioenergetics. CL is involved in the mitochondria-dependent pathway of apoptosis, participates in the function and stabilization of mitochondrial respiratory complexes and supercomplexes and also contributes to mitochondrial integrity. Contributed by Sparagna G 2016-04-18 |
| Cardiovascular Exercise Research Group | CERG |
The Cardiovascular Exercise Research Group (CERG) was established in January 2008 and their research focuses on identifying the key cellular and molecular mechanisms underlying the beneficial effects of physical exercise on the heart, arteries and skeletal muscle in the context of disease prevention and management through experimental, clinical and epidemiological studies. Since 2003 this research group organizes the biennial seminar "Exercise in Medicine" in Trondheim, Norway. |
| Carnitine | Car | Carnitine is an important factor for the transport of long-chain fatty acids bound to carnitine (carnitine acyltransferase) into the mitochondrial matrix for subsequent β-oxidation. There are two enantiomers: D- and L-carnitine. Only the L-isomer is physiologically active. |
| Carnitine O-octanoyltransferase | COT | Carnitine O-octanoyltransferase is a mitochondrial enzyme that transfers carnitine to octanoyl-CoA to form Coenzyme A and octanoylcarnitine: Octanoyl-CoA + L-carnitine ↔ CoA + L-octanoylcarnitine. |
| Carnitine acetyltransferase | CrAT | Carnitine acetyltransferase (CrAT) is located in the mitochondrial matrix and catalyses the formation of acetyl-carnitine from acetyl-CoA and L-carnitine and thus regulates the acetyl-CoA/free CoA ratio which is essential for pyruvate dehydrogenase complex (PDC) activity. |
| Carnitine acyltransferase | Carnitine acyltransferases mediate the transport of long-chain fatty acids across the inner mt-membrane by binding them to carnitine. First, long-chain fatty acids are activated by an energy-requiring step in which the fatty acid ester of CoA is formed enzymatically at the expense of ATP. The fatty acids then pass through the inner mt-membrane and enter the mitochondria as carnitine esters (acylcarnitines). The fatty acyl group is then transferred from carnitine to intramitochondrial CoA and the resulting fatty acyl CoA is used as a substrate in the fatty acid oxidation (FAO) cycle in the mt-matrix. | |
| Carnitine palmitoyltransferase I | CPT-I | Carnitine palmitoyltransferase I (CPT-I, also known as carnitine acyltransferase I) is a regulatory enzyme in mitochondrial long-chain acyl-CoA uptake and further oxidation. CPT-I is associated with the mt-outer membrane mtOM and catalyses the formation of acylcarnitines from acyl-CoA and L-carnitine. In the next step, acyl-carnitines are transported to the mitochondrial matrix via carnitine-acylcarnitine translocase in exchange for free carnitine. In the inner side of the mtIM carnitine palmitoyltransferase II converts the acyl-carnitines to carnitine and acyl-CoAs. There are three enzyme isoforms: CPT-1A (liver type), CPT-1B (muscle type), CPT-1C (brain type). Isoforms have significantly different kinetic and regulatory properties. Malonyl-CoA is an endogenous inhibitor of CPT-I. |
| Carnitine palmitoyltransferase II | CPT-II | Carnitine palmitoyltransferase II (CPT-II, also known as carnitine acyltransferase II) is part of the carnitine shuttle which is responsible for the mitochondrial transport of long-chain fatty acids. CPT-II is located on the inner side of the mtIM and converts the acylcarnitines (produced in the reaction catalyzed by carnitine palmitoyltransferase I) to carnitine and acyl-CoAs, which undergo ß-oxidation in the mitochondrial matrix. Free carnitines are transported out of the mitochondrial matrix in exchange for acyl-carnitines via an integral mtIM protein carnitine-acylcarnitine translocase (CACT). Short- and medium-chain fatty acids do not require the carnitine shuttle for mitochondrial transport. |
| Carnitine-acylcarnitine translocase | CACT | Carnitine-acylcarnitine translocase (CACT) is part of the carnitine shuttle which mediates the mitochondrial transport of long-chain fatty acids where the fatty acid oxidation occurs. CACT is an internal mt-IM protein and transports acylcarnitines into the mitochondrial matrix in exchange for free carnitine. |
| Carrier control titrations | Most of the nonpolar compounds have to be diluted in organic solvents such as DMSO or acetonitrile in order to use them for the titrations in the SUIT protocols. However, the solvent (carrier) itself could affect the mitochondrial physiology and promote alterations that we need to take into account. For this reason, it is necessary to run in parallel to our treatment experiment a control experiment on which we will add a carrier control titration to test if it affects our sample or not. | |
| Catalase | Ctl | Catalase catalyzes the dismutation of hydrogen peroxide to water and oxygen. Perhaps all cells have catalase, but mitochondria of most cells lack catalase. Cardiac mitochondria are exceptional in having mt-catalase activity (rat heart mitochondria: Radi et al 1991; mouse heart mitochondria: Rindler et al 2013). Hydroxylamine is an inhibitor of catalase, which is also inhibited by cyanide and azide. Mitochondrial respiration medium MiR05 was developed considering the intracellular conditions of mitochondria in living cells. In mitochondrial preparations, enzymes and substrates present in the cytosol (such as catalase) are diluted when the plasma membrane is removed. Therefore, the addition of catalase is recommended when working with mitochondrial preparations, to consume any H2O2 generated during the assay. |
| Catalytic activity | kat | Catalytic activity of an enzyme is measured by an enzyme assay and is expressed in units of katal (kat [mol∙s-1]). More commonly (but not conforming to SI units or IUPAC recommendations) enzyme activity is expressed in units U [mol∙min-1]. |
| Cataplerosis | Cataplerosis is the exit of TCA cycle intermediates from the mt-matrix space. | |
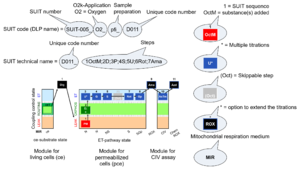
| Categories of SUIT protocols | SUIT-catg |
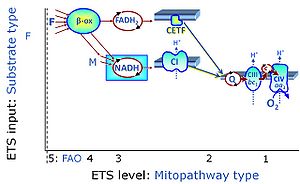
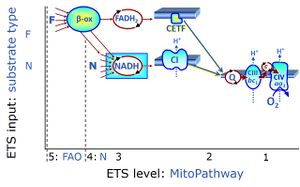
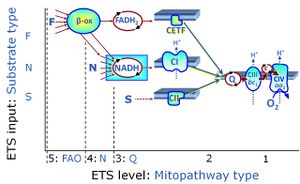
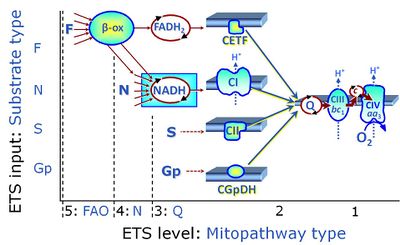
Categories of SUIT protocols group SUIT protocols according to all substrate types involved in a protocol (F, N, S, Gp), independent of the sequence of titrations of substrates and inhibitors which define the Electron-transfer-pathway states. The N-type substrates are listed in parentheses, independent of the sequence of titrations. ROX states may or may not be included in a SUIT protocol, which does not change its category. Similarly, the CIV assay may or may not be added at the end of a SUIT protocol, without effect on the category of a SUIT protocol.
|
| Cell Symposia |  Organized by the editors of Cell Press's leading journals, Cell Symposia bring together exceptional speakers and scientists to discuss topics at the forefront of scientific research. Organized by the editors of Cell Press's leading journals, Cell Symposia bring together exceptional speakers and scientists to discuss topics at the forefront of scientific research. | |
| Cell count and normalization in HRR | Nce | The cell count Nce is the number of cells, expressed in the abstract unit [x] (1 Mx = 106 x). The elementary entity cell Uce [x] is the real unit, the 'single individual cell'. A cell count is the multitude or number N of cells, Nce = N·Uce (Gnaiger MitoFit Preprints 2020.4). Normalization of respiratory rate by cell count yields oxygen flow IO2 expressed in units [amol·s-1·x-1] (=10-18 mol·s-1·x-1). |
| Cell culture media | Cell culture media, like RPMI or DMEM, used for HRR of living cells. | |
| Cell respiration | Cell respiration channels metabolic fuels into the chemiosmotic coupling (bioenergetic) machinery of oxidative phosphorylation, being regulated by and regulating oxygen consumption (or consumption of an alternative final electron acceptor) and molecular redox states, ion gradients, mitochondrial (or microbial) membrane potential, the phosphorylation state of the ATP system, and heat dissipation in response to intrinsic and extrinsic energy demands. See also respirometry. In internal or cell respiration in contrast to fermentation, redox balance is maintained by external electron acceptors, transported into the cell from the environment. The chemical potential between electron donors and electron acceptors drives the electron transfer pathway, generating a chemiosmotic potential that in turn drives ATP synthesis. | |
| Cellular substrates | Ce; Cm | (1) Cellular substrates in vivo, endogenous; Ce.
(2) Cellular substrates in vivo, with exogenous substrate supply from culture medium or serum; Cm.
|
| Chamber volume | The chamber volume of the O2k is 2.0 mL or 0.5 mL of aqueous medium with or without sample, excluding the volume of the stirrer and the volume of the capillary of the stopper (see: Cell count and normalization in HRR). A modular extension of the O2k, the O2k-sV-Module, was specifically developed to perform high-resolution respirometry with reduced amounts of biological sample, and all components necessary for the smaller operation volume of 0.5 mL. | |
| Channel | F7 | » See O2k signals and output |
| Charge | Qel [C] | Charge Qel is the quantity of electricity expressed in the SI unit coulomb [C]. QelX [C] indicates the charge carried by the quantity of a specified ion X. |
| Charge number | zX | The charge number of an ion X or electrochemical reaction with unit stoichiometric number of X is the particle charge [C·x-1] divided by the elementary charge [C·x-1]. The particle charge QNX is the charge per count of ions X or per ion X transferred in the reaction as defined in the reaction equation. |
| Check for updates - DatLab | Check for updates in the Help pull-down menu of DatLab 8 and follow the simple installation instruction if your computer (Linux or Windows) running DatLab is connected to the internet. Alternatively, use a different computer connected to the internet, download the update for Linux, and transfer it to the computer operating DatLab by USB. Check regularly for updates. | |
| Chemical background | CHB, Chb |  Chemical background Chb is due to autooxidation of the reagents. During CIV assays, ascorbate and TMPD are added to maintain cytochrome c in a reduced state. External cytochrome c may be included in the CIV assay. The autooxidation of these compounds is linearly oxygen-dependent down to approximately 50 µM oxygen and responsible for the chemical background oxygen flux after the inhibition of CIV. Oxygen flux due to the chemical reaction of autooxidation must be corrected for the instrumental O2 background. The correction for chemical background is necessary to determine CIV activity, in which case the instrumental O2 background and chemical background may be combined in an overall correction term. Chemical background Chb is due to autooxidation of the reagents. During CIV assays, ascorbate and TMPD are added to maintain cytochrome c in a reduced state. External cytochrome c may be included in the CIV assay. The autooxidation of these compounds is linearly oxygen-dependent down to approximately 50 µM oxygen and responsible for the chemical background oxygen flux after the inhibition of CIV. Oxygen flux due to the chemical reaction of autooxidation must be corrected for the instrumental O2 background. The correction for chemical background is necessary to determine CIV activity, in which case the instrumental O2 background and chemical background may be combined in an overall correction term. |
| Chemical potential | µB [J/mol] | The chemical potential of a substance B, µB [J/mol], is the partial derivative of Gibbs energy, G [J], per amount of B, nB [mol], at constant temperature, pressure, and composition other than that of B,
µB = (∂G/∂nB)T,p,nj≠B The chemical potential of a solute in solution is the sum of the standard chemical potential under defined standard conditions and a concentration (activity)-dependent term, µB = µB° + RT ln(aB)The standard state for the solute is refered to ideal behaviour at standard concentration, c° = 1 mol/L, exhibiting infinitely diluted solution behaviour [1]. µB° equals the standard molar Gibbs energy of formation, ΔfGB° [kJ·mol-1]. The formation process of B is the transformation of the pure constituent elements to one mole of substance B, with all substances in their standard state (the most stable form of the element at 100 kPa (1 bar) at the specified temperature) [2]. |
| Chinese Society of Mitochondrial Research and Medicine | Chinese-Mit | The Chinese Society of Mitochondrial Research and Medicine (Chinese-Mit) is a member of ASMRM. |
| Chinese numerals | Chinese numerals
The Arabic numeral system used today in China was introduced to China by the Europeans in the early 17th century. But the Chinese character-based number systems are still in use. The financial numerals are used only when writing an amount on a form for remitting money at a bank. They function as anti-fraud numerals. The character 零 (zero) appeared very early in ancient Chinese writing. However, at that time, it did not mean "nothing", but "bits and pieces", "not much". 一百零五(105) means in Chinese: In addition to a hundred, there is a fraction of five. With the introduction of the Arabic numerals, 105 is exactly pronounced “one hundred zero five”, the character 零 corresponds exactly to the symbol 0. Thus, the character 零has the meaning of 0. But the character 〇 was one of the Chinese characters created and promulgated by the only empress (with greater achievements than countless emperors) in the history of China in 690 AD (much later than the invention of 0 in India) for the purpose of demonstrating her power. At that time the character 〇 meant “star”, representing a round planet. It is now used as a synonym for the 零 (zero). | |
| Chloroplasts | pt? | Chloroplasts (Greek chloros: green; plastes: the one who forms) are small structures within the cells that conduct photosynthesis. They are a type of organelle called plastids that are present in eukaryotic plant cells (algae, aquatic and terrestrial plants) and characterized by having two membranes and a high concentration of the pigment Chlorophyll. Like mitochondria, they originated through the endosymbiosis of a cyanobacteria by an early eukaryotic cell and they have their own DNA which replicates during cell division. In addition to photosynthesis, in their internal matrix called stroma they also carry out other metabolic functions within the plant cells such as fatty acid synthesis or amino acid synthesis. |
| Chlororespiration | In chlororespiration oxygen is consumed by a putative respiratory electron transfer system (ETS) within the thylakoid membrane of the chloroplasts and ATP is produced. It is a process that involves the interaction with the photosynthetic ETS in which NAD(P)H dehydrogenase transfers electrons to oxygen with the assistance of the photosynthetic plastoquinone (PQ), which acts as a non-photochemical redox carrier. Initially described in the unicellular alga Chlamydomonas reindhartdii, chlororespiration was highly disputed for years until the discovery of a NAD(P)H-dehydrogenase (NDH) complex (plastidic encoded) and plastid terminal oxidase (PTOX) (nuclear encoded) in higher-plant chloroplasts. PTOX is homologous to the plant mitochondrial alternative oxidase and has the role of preventing the over-reduction of the PQ pool while the NDH complexes provide a gateway for the electrons to form the ETS and consume oxygen. As a result of this process there is a cyclic electron flow around Photosystem I (PSI) that is activated under stress conditions acting as a photoprotection mechanism and could be involved in protecting against oxidative stress. | |
| Choline dehydrogenase | Choline dehydrogenase (EC 1.1.99.1) is bound to the inner mt-membrane, oxidizes choline in kidney and liver mitochondria, with electron transfer into the Q-junction, and is thus part of the Electron transfer pathway. Analogous to succinate dehydrogenase (CII), electron transfer from choline dehydrogenase is FAD-linked downstream to Q. Choline is an ET-pathway substrate types 3. | |
| Citrate | citrate, C6H5O7-3, is a tricarboxylic acid trianion, intermediate of the TCA cycle, obtained by deprotonation of the three carboxy groups of citric acid. Citrate is formed from oxaloacetate and acetyl-CoA through the catalytic activity of the citrate synthase. In the TCA cycle, citrate forms isocitrate by the activity of the aconitase. Citrate can be transported out of the mitochondria by the tricarboxylate transport, situated in the inner mitochondrial membrane. The transport occurs as an antiport of malate from the cytosol and it is a key process for fatty acid and oxaloacetate synthesis in the cytosol. | |
| Citrate synthase | CS | Condensation of oxaloacetate with acetyl-CoA yields citrate as an entry into the TCA cycle. CS is located in the mt-matrix. CS activity is frequently used as a functional marker of the amount of mitochondria (mitochondrial elementary marker, mtE) for normalization of respiratory flux. |
| Citreoviridin | Citreoviridin is an inhibitor of the ATP synthase which, differently from the FO subunit binding inhibitor oligmycin, binds to the F1 subunit of the ATP synthase. | |
| Close and delete file - DatLab | Close and delete file. The decision to delete a DatLab file containing no useful data can be made most easily when viewing the traces. Only available when disconnected from the O2k. | |
| Closed chamber | C | The O2k-chamber can be used as a closed system or open system. Gas bubbles must be avoided. |
| Closed system | A closed system is a system with boundaries that allow external exchange of energy (heat and work), but do not allow exchange of matter. A limiting case is light and electrons which cross the system boundary when work is exchanged in the form of light or electric energy. If the surroundings are maintained at constant temperature, and heat exchange is rapid to prevent the generation of thermal gradients, then the closed system is isothermal. A frequently considered case are closed isothermal systems at constant pressure (and constant volume with aqueous solutions). Changes of closed systems can be partitioned according to internal and external sources. Closed systems may be homogenous (well mixed and isothermal), continuous with gradients, or discontinuous with compartments (heterogenous). | |
| Coenzyme | A coenzyme or cosubstrate is a cofactor that is attached loosely and transiently to an enzyme, in contrast to a prosthetic group that is attached permanently and tightly. The coenzyme is required by the corresponding enzyme for its activity (IUPAC definition). A coenzyme is 'a low-molecular-weight, non-protein organic compound participating in enzymatic reactions as dissociable acceptor or donor of chemical groups or electrons' (IUPAC definition). | |
| Coenzyme A | CoA | Coenzyme A is a coenzyme playing an essential role in the tricarboxylic acid cycle (oxidation of pyruvate to acetyl-CoA) and fatty acid oxidation. CoA is a thiol that reacts with carboxylic acids to form CoA-activated thioesters. |
| Coenzyme Q | Q, CoQ | Coenzyme Q or ubiquinone (2,3-dimethoxy-5-methyl-6-polyprenyl-1,4-benzoquinone) was discovered in 1957 by the group of Crane. It is a lipid composed of a benzoquinone ring with an isoprenoid side chain, two methoxy groups and one methyl group. The length of the isoprenoid chain varies depending on the species; for example, six isoprenoid units (CoQ6) is the most commonly found CoQ in Saccharomyces cerevisiae, eight units in Escherichia coli (CoQ8), nine units in Caenorhabditis elegans and rodents (CoQ9), ten units in humans (CoQ10), and some species have more than one CoQ form, e.g. human and rodent mitochondria contain different proportions of CoQ9 and CoQ10. These redox compounds exist in three different forms: quinone (oxidized), quinol (reduced), and an intermediate semiquinone. More details » Q-junction |
| Coenzyme Q2 | CoQ2 | Coenzyme Q2 or ubiquinone-2 (CoQ2) is a quinone derivate composed of a benzoquinone ring with an isoprenoid side chain consisting of two isoprenoid groups, with two methoxy groups, and with one methyl group. In HRR it is used as a Q-mimetic to detect the redox changes of coenzyme Q at the Q-junction in conjunction with the Q-Module, since the naturally occurring long-chain coenzyme Q (e.g. CoQ10) is trapped within membrane boundaries. CoQ2 can react both with mitochondrial complexes (e.g. CI, CII and CIII) at their quinone-binding sites and with the detecting electrode. |
| Cofactor | A cofactor is 'an organic molecule or ion (usually a metal ion) that is required by an enzyme for its activity. It may be attached either loosely (coenzyme) or tightly (prosthetic group)' (IUPAC definition). | |
| Cole 2000 BMJ | ||
| Comma for separating a term and its abbreviation | , | Should we used a comma for separating a term and its abbreviation in the text? The SI Brochure frequently does not use a comma. The comma might be added, if it helps to clarify the distinction between the term and its abbreviation. The example “reduced Q fraction, Qr” – the sequence of Q and Qr may be confusing without comma. There will always be examples, where it is not clear, if a comma is needed. |
| Communication - mitochondria and the patient | Mitochondria and the patient: communication between patients, medical professionals, scientists, and the public | |
| Comorbidity | Comorbidities are common in obesogenic lifestyle-induced early aging. These are preventable, non-communicable diseases with strong associations to obesity. In many studies, cause and effect in the sequence of onset of comorbidities remain elusive. Chronic degenerative diseases are commonly obesity-induced. The search for the link between obesity and the etiology of diverse preventable diseases lead to the hypothesis, that mitochondrial dysfunction is the common mechanism, summarized in the term 'mitObesity'. | |
| Company of Scientists | CompaSci | The Company of Scientists evolves as a concept for implementing scientific innovations on the market. |
| Comparison of respirometric methods | The comparison of respirometric methods provides the basis to evaluate different instrumental platforms and different mitochondrial preparations, as a guide to select the best approach and to critically evaluate published results. | |
| Complex I | CI | Complex I, NADH:ubiquinone oxidoreductase (EC 1.6.5.3), is an enzyme complex of the Electron transfer pathway, a proton pump across the inner mt-membrane, responsible for electron transfer to ubiquinone from NADH formed in the mt-matrix. CI forms a supercomplex with Complex III. There is a widespread ambiguity on the 'lonely H+ (the lonely hydron)' surrounding Complex I: CI ambiguities. |
| Complex I&II-linked substrate state | NS | See NS-pathway control state (previous: CI&II-linked) |
| Complex I-linked substrate state | N | See N-pathway control state (previous: CI-linked) versus Complex I |
| Complex II | CII | Complex II or succinate:quinone oxidoreductase (SQR) is the only membrane-bound enzyme in the TCA cycle and is part of the electron transfer pathway. The reversible oxidoreduction of succinate and fumarate is catalyzed in a soluble domain and coupled to the reversible oxidoreduction of quinol and quinone in the mitochondrial inner membrane. CII consists in most species of four subunits. The flavoprotein succinate dehydrogenase is the largest polypeptide of CII, located on the matrix face of the mt-inner membrane. Succinate:quinone oxidoreductases (SQRs, SDHABCD) favour oxidation of succinate and reduction of quinone in the canonical forward direction of the TCA cycle and electron transfer into the Q-junction. In contrast, quinol:fumarate reductases (QFRs, fumarate reductases, FRDABCD) tend to operate in the reverse direction reducing fumarate and oxidizing quinol. |
| Complex II ambiguities | CII ambiguities | The current narrative that the reduced coenzymes NADH and FADH2 feed electrons from the tricarboxylic acid (TCA) cycle into the mitochondrial electron transfer system can create ambiguities around respiratory Complex CII. Succinate dehydrogenase or CII reduces FAD to FADH2 in the canonical forward TCA cycle. However, some graphical representations of the membrane-bound electron transfer system (ETS) depict CII as the site of oxidation of FADH2. This leads to the false believe that FADH2 generated by electron transferring flavoprotein (CETF) in fatty acid oxidation and mitochondrial glycerophosphate dehydrogenase (CGpDH) feeds electrons into the ETS through CII. In reality, NADH and succinate produced in the TCA cycle are the substrates of Complexes CI and CII, respectively, and the reduced flavin groups FMNH2 and FADH2 are downstream products of CI and CII, respectively, carrying electrons from CI and CII into the Q-junction. Similarly, CETF and CGpDH feed electrons into the Q-junction but not through CII. The ambiguities surrounding Complex II in the literature call for quality control, to secure scientific standards in current communications on bioenergetics and support adequate clinical applications. |
| Complex II-linked substrate state | SRot, S | See S-pathway control state (previous: CII-linked) |
| Complex III | CIII | Complex III or coenzyme Q : cytochrome c - oxidoreductase, sometimes also called the cytochrome bc1 complex is a complex of the electron transfer pathway. It catalyzes the reduction of cytochrome c by oxidation of coenzyme Q (CoQ) and the concomitant pumping of 4 protons from the cathodic (negative) mitochondrial matrix to the anodic (positive) intermembrane space. |
| Complex IV | CIV | Complex IV or cytochrome c oxidase is the terminal oxidase of the mitochondrial electron transfer system, reducing oxygen to water, with reduced cytochrome c as a substrate. Concomitantly to that, CIV pumps protons against the electrochemical protonmotive force. CIV is frequently abbreviated as COX or CcO. It is the 'ferment' (Atmungsferment) of Otto Warburg, shown to be related to the cytochromes discovered by David Keilin. |
| Concentration | c [mol·L-1]; C [x·L-1] | Concentration [mol·L-1] is a volume-specific quantity for diluted samples s. In a concentration, the sample is expressed in a variety of formats: count, amount, charge, mass, energy. In solution chemistry, amount concentration is amount of substance nB per volume V of the solution, cB = [B] = nB·V-1 [mol·dm-3] = [mol·L-1]. The standard concentration, c°, is defined as 1 mol·L-1 = 1 M. Count concentration CX = NX·V-1 [x·L-1] is the concentration of the number NX of elementary entities X, for which the less appropriate term 'number concentration' is used by IUPAC. If the sample is expressed as volume Vs (e.g., VO2), then the 'volume-concentration' of Vs in V is termed 'volume fraction', Φs = Vs·V-1 (e.g., volume fraction of O2 in dry air, ΦO2) = 0.20946). Density is the mass concentration in a volume VS of pure sample S. A change of concentration, dcX, in isolated or closed systems at constant volume is due to internal transformations (advancement per volume) only. In closed compressible systems (with a gas phase), the concentration of the gas changes, when pressure-volume work is performed on the system. In open systems, a change of concentration can additionally be due to external flow across the system boundaries. |
| Conflict of interest | As stated on the Bioenergetics Communications' policy, a conflict of interest may be of non-financial or financial nature. Examples of conflicts of interest include (but are not limited to):
| |
| Connect to O2k | Connect to O2k connects DatLab with the O2k. Select the USB port (or Serial port) with the corresponding cable connecting your PC to the O2k. Select the subdirectory for saving the DLD file. Then data recording starts with experimental time set at zero. | |
| Connection window | After starting DatLab either the Connection window opens automatically by default or open O2k control by pressing [F7] and select the communication port. | |
| Convergent electron flow | n.a. | Convergent electron flow is built into the metabolic design of the Electron transfer pathway. The glycolytic pathways are characterized by important divergent branchpoints: phosphoenolpyruvate (PEPCK) branchpoint to pyruvate or oxaloactetate; pyruvate branchpoint to (aerobic) acetyl-CoA or (anaerobic) lactate or alanine. The mitochondrial Electron transfer pathway, in contrast, is characterized by convergent junctions: (1) the N-junction and F-junction in the mitochondrial matrix at ET-pathway level 4, with dehydrogenases (including the TCA cycle) and ß-oxidation generating NADH and FADH2 as substrates for Complex I and electron-transferring flavoprotein complex, respectively, and (2) the Q-junction with inner mt-membrane respiratory complexes at ET-pathway level 3, reducing the oxidized ubiquinone and partially reduced semiquinone to the fully reduced ubiquinol, feeding electrons into Complex III. |
| Copy marks | In Copy marks, Marks in DatLab are copied from a seleted Plot to the active plot. | |
| Copy marks - DatLab | ||
| Copy to clipboard | In DatLab Copy to clipboard can be used to copy selected graphs or values and to paste them to your preferred program or file (e.g. Word, Excel). | |
| Copyright | Authors retain the copyright for the contents of their manuscripts published in Bioenergetics Communications. {Quote} All preprints are posted with a Creative Commons CC BY 4.0 license, ensuring that authors retain copyright and receive credit for their work, while allowing anyone to read and reuse their work. {end of Quote} | |
| Count | NX [x] | Count NX is the number N of elementary entities of entity-type X. The single elementary entity UX is a countable object or event. NX is the number of objects of type X, whereas the term 'entity' and symbol X are frequently used and understood in dual-message code indicating both (1) the entity-type X and (2) a count of NX = 1 x for a single elementary entity UX. 'Count' is synonymous with 'number of entities' (number of particles such as molecules, or objects such as cells). Count is one of the most fundamental quantities in all areas of physics to biology, sociology, economy and philosophy, including all perspectives of the statics of countable objects to the dynamics of countable events. The term 'number of entities' can be used in short for 'number of elementary entities', since only elementary entities can be counted, and as long as it is clear from the context, that it is not the number of different entity types that are the object of the count. |
| Coupled respiration | Coupled respiration drives oxidative phosphorylation of the diphosphate ADP to the triphosphate ATP, mediated by proton pumps across the inner mitochondrial membrane. Intrinsically uncoupled respiration, in contrast, does not lead to phosphorylation of ADP, despite of protons being pumped across the inner mt-membrane. Coupled respiration, therefore, is the coupled part of respiratory oxygen flux that pumps the fraction of protons across the inner mt-membrane which is utilized by the phosphorylation system to produce ATP from ADP and Pi. In the OXPHOS state, mitochondria are in a partially coupled state, and the corresponding coupled respiration is the free OXPHOS capacity. In the state of ROUTINE respiration, coupled respiration is the free ROUTINE activity. | |
| Coupling-control efficiency | Coupling-control efficiencies are flux control efficiencies jZ-Y at a constant ET-pathway competent state. | |
| Coupling-control protocol | CCP | A coupling-control protocol CCP induces different coupling control states at a constant electron-transfer-pathway state. Residual oxygen consumption (Rox) is finally evaluated for Rox correction of flux. The CCP may be extended, when further respiratory states (e.g. cell viability test; CIV assay) are added to the coupling control module consisting of three coupling control states. The term phosphorylation control protocol, PCP, has been introduced synonymous for CCP. » MiPNet article |
| Coupling-control ratio | CCR | Coupling-control ratios CCR are flux control ratios FCR at a constant mitochondrial pathway-control state. In mitochondrial preparations, there are three well-defined coupling states of respiration: LEAK respiration, OXPHOS, and Electron-transfer-pathway state (ET state). In these states, the corresponding respirtory rates are symbolized as L, P, and E. In living cells, the OXPHOS state cannot be induced, but in the ROUTINE state the respiration rate is R. A reference rate Z is defined by taking Z as the maximum flux, i.e. flux E in the ET-state, such that the lower and upper limits of the CCR are defined as 0.0 and 1.0. Then there are two mitochondrial CCR, L/E and P/E, and two CCR for living cells, L/E and R/E. |
| Coupling-control state | CCS | Coupling-control states are defined in mitochondrial preparations (isolated mitochondria, permeabilized cells, permeabilized tissues, homogenates) as LEAK respiration, OXPHOS, and ET states, with corresponding respiration rates (L, P, E) in any electron-transfer-pathway state which is competent for electron transfer. These coupling states are induced by titration of ADP and uncouplers, and application of specific inhibitors of the phosphorylation pathway. In living cells, the coupling-control states are LEAK respiration, ROUTINE, and ET states of respiration with corresponding rates L, R, E, using membrane-permeable inhibitors of the phosphorylation system (e.g. oligomycin) and uncouplers (e.g. CCCP). Coupling-control protocols induce these coupling-control states sequentially at a constant electron-transfer-pathway state. |
| Coupling/pathway control diagram | CPCD | Coupling/pathway control diagrams illustrate the respiratory states obtained step-by-step in substrate-uncoupler-inhibitor titrations in a SUIT protocol. Each step (to the next state) is defined by an initial state and a metabolic control variable, X. The respiratory states are shown by boxes. X is usually the titrated substance in a SUIT protocol. If X (ADP, uncouplers, or inhibitors of the phosphorylation system, e.g. oligomycin) exerts coupling control, then a transition is induced between two coupling-control states. If X (fuel substrates, e.g. pyruvate and succinate, or Electron transfer pathway inhibitors, e.g. rotenone) exerts pathway control, then a transition is induced between two Electron-transfer-pathway states. The type of metabolic control (X) is shown by arrows linking two respiratory states, with vertical arrows indicating coupling control, and horizontal arrows indicating pathway control. Marks define the section of an experimental trace in a given respiratory state (steady state). Events define the titration of X inducing a transition in the SUIT protocol. The specific sequence of coupling control and pathway control steps defines the SUIT protocol pattern. The coupling/pathway control diagrams define the categories of SUIT protocols (see Figure). |
| Cover-Slip\black | A Cover-Slip should be placed on top of the O2k-Stopper to minimize contamination and evaporation of liquid extruding from the capillary of the stopper. The Cover-Slips do not exert any direct effect on oxygen backdiffusion into the O2k-chamber. Use the the Cover-Slip\black to avoid light penetration and disturbance of fluorescence signals and generally for optical measurements in the O2k. | |
| Crabtree effect | The Crabtree effect describes the observation that respiration is frequently inhibited when high concentrations of glucose or fructose are added to the culture medium - a phenomenon observed in numerous cell types, particularly in proliferating cells, not only tumor cells but also bacteria and yeast. The Pasteur effect (suppression of glycolysis by oxygen) is the converse of the Crabtree effect (suppression of respiration by high concentration of glucose or fructose). | |
| Creatine | Cr | Creatine is a nitrogenous organic acid that occurs naturally in vertebrates and helps primarily muscle cells to supply energy by increasing the formation of adenosine triphosphate (ATP). |
| Creatine kinase | CK | The mitochondrial creatine kinase, also known as phosphocreatine kinase (CPK), facilitates energy transport with creatine and phosphocreatine as diffusible intermediates. |
| Creative Commons Attribution License | Open Access preprints (not peer-reviewed) and articles (peer-reviewed) distributed under the terms of the Creative Commons Attribution License allow unrestricted use, distribution, and reproduction in any medium, provided the original authors and source are credited. © remains with the authors, who have granted the publisher license in perpetuity. | |
| Critical oxygen pressure | pc | The critical oxygen pressure, pc, is defined as the partial oxygen pressure, pO2, below which aerobic catabolism (respiration or oxygen consumption) declines significantly. If anaerobic catabolism is activated simultaneously to compensate for lower aerobic ATP generation, then the limiting oxygen pressure, pl, is equal to the pc. In many cases, however, the pl is substantially lower than the pc. |
| Cross-linked respiratory states | CLRS | Coordinated respiratory SUIT protocols are designed to include cross-linked respiratory states, which are common to these protocols. Different SUIT protocols address a variety of respiratory control steps which cannot be accomodated in a single protocol. Cross-linked respiratory states are included in each individual coordinated protocol, such that these states can be considered as replicate measurements, which also allow for harmonization of data obtained with these different protocols. |
| Curcumin | Curcumin has been shown to possess significant anti-inflammatory, anti-oxidant, anti-carcinogenic, anti-mutagenic, anti-coagulant and anti-infective effects. The protective effects of curcumin on rat heart mitochondrial injuries induced by in vitro anoxia–reoxygenation were evaluated by Xu et al 2013. It was found that curcumin added before anoxia or immediately prior to reoxygenation exhibited remarkable protective effects against anoxia–reoxygenation induced oxidative damage to mitochondria. | |
| Custom label | A Custom label can be entered in this box to rename the axis label. Two lines are available for the axis name and unit. | |
| Custom-made stoppers | Stoppers can be custom-made for applications with user-specific sensors according to customer specifications. | |
| Cuvettes | Cuvettes are used in fluorometry and transmission spectrophotometry to contain the samples. Use of the term 'cells' for cuvettes is discouraged, to avoid confusion with 'living cells'. Traditionally cuvettes have a square cross-section (10 x 10 mm). For many applications they are made of transparent plastic. Glass cells are used where samples may contain plastic solvents, and for some applications requiring measurements below 300 nm, quartz glass or high purity fused silica cuvettes may be necessary. | |
| Cyanide | KCN | Cyanide (usually added as KCN) is a competitive inhibitor of cytochcrome c oxidase (CIV). Inhibition is reversed by pyruvate and high oxygen levels. |
| Cyclic voltammetry | CV | Cyclic voltammetry (CV) is a type of electrochemical measurement which is applied with the Q-Module as quality control to
(1) determine the oxidation and reduction peak potentials of Coenzyme Q in the specific experimental condition, (2) check the quality of the Q-Sensor, and (3) test the interference of chemicals used in the HRR assay with the Q-Sensor. In CV, the Q-Sensor with the three-electrode system is used to obtain information about the analyte (CoQ) by measuring the current (I) as the electric potential (V) between two of the electrodes is varied. In CV the electric potential between the glassy carbon (GC) and the Ag/AgCl reference electrode changes linearly versus time in cyclical phases, while the current is detected between GC and platinum electrode (Pt). The detected current is plotted versus the applied voltage to obtain the typical cyclic voltammogram trace (Figure 1). The presence of substances that are oxidized/reduced will result in current between GC and Pt, which can be seen as characteristic peaks in the voltammogram at a defined potential. The oxidation or the reduction peak potential values are used to set the GC (integrated into the Q-Sensor) for a separate experiment to measure the Q redox state of a biological sample. The oxidation and reduction peak potentials can be influenced by 1) the respiration medium, 2) the type of CoQ, 3) the polarization window, 4) the scan speed, 5) the number of cycles, 6) the concentration of the analyte (CoQ), and 7) the initial polarization voltage. <be> |
| Cyclic voltammetry - DatLab | Cyclic voltammetry | |
| Cyclosporin A | CsA | Cyclosporin A (CsA) is a cyclic undecapeptide from an extract of soil fungi that binds the cyclophilin D and thus preventing the formation of the mitochondrial permeability transition pore. The interaction of CsA with the cyclophilin D is phosphate mediated but the full mechanism of interaction is not well understood. For example, the deficiency of cyclophilin D in KO models does not prevent mitochondria from permeability transition and from CsA inhibition. Moreover, it is also a is a calcineurin inhibitor and potent immunosuppressive agent used largely as a means of prophylaxis against cellular rejection after solid organ transplantation. |
| Cytochrome c | c | Cytochrome c is a component of the Electron transfer-pathway (Electron transfer pathway) in mitochondria. It is a small heme protein loosely associated with the outer side of the inner mitochondrial membrane. The heme group of cytochrome c transfers electrons from Complex III to Complex IV. The release of cytochrome c into the cytoplasm is associated with apoptosis. Cytochrome c is applied in HRR to test the integrity of the mitochondrial outer membrane (cytochrome c control efficiency). |
| Cytochrome c control efficiency | jcyt c | The cytochrome c control efficiency expresses the control of respiration by externally added cytochrome c, c, as a fractional change of flux from substrate state CHNO to CHNOc. These fluxes are corrected for Rox and may be measured in the OXPHOS state or ET state, but not in the LEAK state. In this flux control efficiency, CHNOc is the reference state with stimulated flux; CHNO is the background state with CHNO substrates, upon which c is added: jcyt c = (JCHNOc-JCHNO)/JCHNOc. |
| D-number | D### | D number is the unique code given for each SUIT protocol. In the same SUIT protocol family (SUIT-###), there might be different protocols, specifically designed for different sample type (e.g., different mitochondrial preparations) or for different applications (e.g., O2, AmR, Fluo, MgG). Since the use of different kinds of sample or application may result in slightly different steps, each protocol receives a different D-number. |
| DORA | DORA | The Declaration on Research Assessment DORA recognizes the need to improve the ways in which researchers and the outputs of scholarly research are evaluated. |
| DTPA | DTPA | DTPA (Diethylenetriamine-N,N,N',N,N-pentaacetic acid, pentetic acid,(Carboxymethyl)imino]bis(ethylenenitrilo)-tetra-acetic acid) is a polyaminopolycarboxylic acid (like EDTA) chelator of metal cations. DTPA wraps around a metal ion by forming up to eight bounds, because each COO- group and and N-center serves a center for chelation. With transition metals the number of bounds is less than eight. The compound is not cell membrane permeable. In general, it chelates multivalent ions stronger than EDTA. |
| DatLab | DatLab is the O2k-Software for Data Acquisition & Analysis, specifically developed for high-resolution respirometry with the O2k.
The newest DatLab version is DatLab 8, included in the O2k-Packages. NextGen-O2k and O2k-Series J* and higher come with DatLab 8 installed on the integrated PC (Linux). DatLab 8 is required for the NextGen-O2k. DatLab 8.1 is compatible with O2k-Series (E and higher). The DatLab software is designed for 64-bit versions of Windows operating systems and does not run on MAC devices. The minimum computer requirements are Intel-Core-2 or equivalent CPU, 2GB RAM and Windows XP (64-bit version). However, we recommend Intel i5 or equivalent CPU, 4GB RAM, Windows 10 (64-bit version) and SSD. For the proper display of DatLab on your computer, please make sure the “Language settings” are set to English. *Optionally available without integrated PC. | |
| DatLab 2 | DL2 | DatLab 2 (DL2) is a MS-DOS programe. DL2 is still used for analysis of oxygen kinetics, after exporting files recorded in recent DatLab versions. A user-friendly O2-kinetics module is in preparation (DL8). |
| DatLab and SUIT protocols | This is a brief summary of steps to be taken for performing a high-resolution respirometry experiment with SUIT protocols using the OROBOROS Oroboros O2k and DatLab software. (1) Search for a specific SUIT protocol name (go to MitoPedia: SUIT). The list of MitoPedia SUIT protocols can be sorted by categories of SUIT protocols (sorting by SUIT protocol name), which is listed as the 'abbreviation' of the SUIT protocol name. (2) Copy the template for Mark names into your DatLab subdirectory: DatLab\APPDATA\MTEMPLAT. (3) Copy the DatLab-Analysis template for this SUIT protocol. (4) Follow the link to the corresponding publication or MiPNet communication, where the pdf file describing the SUIT protocol is available. (5) A DatLab demo file may be available providing an experimental example. After each sequential titration, a mark is set on the plot for flux or flow. After having set all marks, pull down the 'Mark names' menu, select the corresponding SUIT protocol for mark names, and rename all marks. The Mark names template also provides standard values of the titration volume preceding each mark. (6) Go to 'Mark statistics' [F2], copy to clipboard, and paste into the sample tab in the DatLab-Analysis template.
| |
| DatLab data file | dld8, DLD | DatLab 8: The file type generated is *.dld8. DatLab 7: The file type generated is *.DLD. |
| DatLab error messages | Common DatLab error messages and according actions and solutions are listed here. | |
| DatLab installation | We recommend a 'clean install' for DatLab installation: rename your previous DatLab programme subdirectory (e.g. C:\DatLab_OLD). The standard Instrumental and SUIT DL-Protocols package is automatically implemented with the simple DatLab programme installation. | |
| DatLab oxygen flux: performance and data analysis | The quality of the results are strongly affected by the performance and data analysis. Therefore, we provide guidelines for performing and evaluating respirometric assays. | |
| DatLab templates | DatLab templates can be imported for O2k-setups, graph layouts, mark names, TIP2k setups and marks statistics configurations.
| |
| DatLab-Analysis templates | Go in DatLab to Mark statistics (F2), select which type of marks you want to export ("All marks in plot" or "DL-Protocol marks", with 3 possibilities each), then click on [Copy to clipboard] to copy selected values and paste them to a DatLab-Analysis template for numerical and graphical data analysis. | |
| DatLab-Upgrading\4.x-5.2 | DatLab-Upgrading\4.1-5.2: Upgrading DatLab 4.x to 5.2, incl. O2k-Manual, with free follow-up updates of DatLab 5.2. Discontinued: see higher DatLab version. | |
| Data labels and units - DatLab | ||
| Data masks - DatLab | ||
| Data recording interval | F7 | The data recording interval is the time interval at which data is sampled with an instrument. In DatLab the data recording interval is set in the O2k control window. The system default value is 2 s. A lower data recording interval is selected for kinetic experiments, and when the volume-specific oxygen flux is high (>300 pmol O2·s-1·ml-1). Technically, the O2k instrument (hardware) measures the sensor signal every 10ms (which is NOT the „data recording interval“). By the given data recording interval from DatLab (software) a discrete number of sensor signal points are taken to calculate an average value in the O2k (e.g. a data recording interval of 2 s can take 200 sensor signal points; a data recording interval of 0.5 s can take 50 sensor signal points). This average value is sent to DatLab and is recorded as a raw data point. However, there is a defined threshold: the O2k does not apply more than 200 sensor signal points to calculate the average for the raw data point. For example a data recording interval of 3 s could take 300 sensor signal points but only the 200 most recent sensor signal points are used for averaging. |
| DataCite | DataCite | DataCite is a global community of organizations and researchers identifying and citing research outputs and resources. We provide services to create persistent records of research, enable discovery and reuse, and support workflows throughout the research lifecycle. |
| Dataset | A dataset is a collection of data. In the context of databases a dataset represents the collection of entries in a database-table. In this table columns represent attributes and rows display the according values of the entries. | |
| De Onis 2007 Bull World Health Organization | ||
| Dead cells | dce | Dead cells dce are characterized by the loss of plasma membrane barrier function. The total cell count (Nce) is the sum of viable cells (Nvce) and dead cells (Ndce). |
| Decimal marker and spaces between groups of numerals | . | A decimal marker is used to separate the integral part of numbers from the decimal part. The SI recommends: "the symbol for the decimal marker shall be either the point on the line or the comma on the line". In English language versions, the dot (point on the line) should be used uniquely as the decimal marker. To avoid ambiguities, BEC follows the SI recommendation that “Numbers may be divided in groups of three in order to facilitate reading; neither dots nor commas are ever inserted in the spaces between groups” (pages 183-184). |
| Default label | The Default label is the system default value for the axis label. These labels are changed automatically, according to the selected channel and unit. To change this label enter a Custom label. | |
| Delete points | Select Delete points in the Mark information window to remove all data points in the marked section of the active plot. See also Interpolate points and Restore points or Recalculate slope. | |
| Density | ρ, C, D | Density, mass density ρ = m·V-1 [kg·m-3], is mass m divided by volume V. Surface density ρA = m·A-1 [kg·m-2] (SI). For a pure sample S, the mass density ρS = mS·VS-1 [kg·m-3] is the mass m of pure sample S per volume VS of the pure sample. With density ρ thus defined, the 'amount density' of substance B is ρB = nB·VB-1 [mol·m-3]. This is not a commonly used expression, but the inverse is defined as the molar volume of a pure substance (IUPAC), Vm,B = VB·nB-1 [m3·mol-1]. The pure sample is a pure gas, pure liquid or pure solid of a defined elementary entity. The amount concentration, cB = nB·V-1 [mol·m-3] is the amount nB of substance B divided by the volume V of the mixture (IUPAC), and this is not called an 'amount density'. The term 'amount density' is reserved for an amount of substance per volume VS of the pure substance. This explicit distinction between 'density' related to the volume of the sample and 'concentration' related to the total volume of the mixture is very helpful to avoid confusion. Further clarification is required in cases, when the mass density ρs of the sample in the mixture differs from the mass density ρS of the pure sample before mixing. Think of a sample S of pure ethanol with a volume of 1 L at 25 °C, which is mixed with a volume of 1 L of pure water at 25 °C: after the temperature of the mixture has equilibrated to 25 °C, the total volume of the mixture is less than 2 L, such that the volume VS of 1 L pure ethanol has diminished to a smaller volume Vs of ethanol in the mixture; the density of ethanol in the mixture is higher than the density of pure ethanol (this is incomplete additivity). The volume Vs of sample s in a mixture is by definition smaller than the total volume V of the mixture. Sample volume VS and system volume V are identical, but this applies only to the case of a pure sample. Concentration is related to samples s per total volume V of the mixture, whereas density is related to samples S or s per volume VS = V or Vs < V, respectively (BEC 2020.1). |
| Derivative spectroscopy | Derivative spectroscopy can be used to eliminate interfering artefacts or species. A first order derivative will remove a constant background absorbance across the spectral range. A second order derivative spectrum will remove a species whose absorbance is linearly dependent upon the wavelength, etc.. | |
| Deselect channels | F7 | Channels can be selected/deselected in DatLab in the O2k configuration. Deselect all O2k-MultiSensor channels in O2k-Core applications. Select only the specifically used channels in O2k-MultiSensor applications. |
| Detector | A detector is a device that converts the light falling upon it into a current or voltage that is proportional to the light intensity. The most common devices in current use for fluorometry and spectrophotometry are photodiodes and photodiode arrays. | |
| Diapause | Diapause is a preprogrammed form of developmental arrest that allows animals to survive harsh environmental conditions and may also allow populations to synchronize periods of growth and reproduction with periods of optimal temperatures and adequate water and food. Diapause is endogenously controlled, and this dormancy typically begins well before conditions become too harsh to support normal growth and development [1,2]. » MiPNet article | |
| Dicarboxylate carrier | DIC | The dicarboxylate carrier is a transporter which catalyses the electroneutral exchange of malate2- (or succinate2-) for inorganic phosphate, HPO42-. |
| Difference spectrum | A difference spectrum is an absorbance spectrum obtained by subtracting that of one substance from that of another. For example, a difference spectrum may be plotted of the absorbance spectrum obtain ed from reduced cytochrome c and subtracting the absorbance spectrum from the same concentration of cytochrome c in its oxidised state. The difference spectrum may be used to quantify the amount to which the cytochrome c is reduced. This can be achieved with the aid of a reference spectrum (or spectra) and the least squares method. | |
| Different O2 fluxes in left and right chamber | What are potential causes for different O2 fluxes in the left and right chamber? | |
| Diffraction gratings | Diffraction gratings are dispersion devices that are made from glass etched with fine grooves, spaced at the same order of magnitude as the wavelength of the light to be dispersed, and then coated with aluminium to reflect the light to the photodiode array. Diffraction gratings reflect the light in different orders and filters need to be incorporated to prevent overlapping. | |
| Digital Object Identifier | DOI | A Digital Object Identifier, DOI, is a persistent identifier used to uniquely identify online publications in order to ensure they remain traceable, searchable and citable over the long term. Compared to other types of persistent identifiers, the DOI system is widespread and well established in the life sciences arena, and it provides widely accepted visible proof that a publication is citable. |
| Digitonin | Dig | Digitonin is a mild detergent that permeabilizes plasma membranes selectively due to their high cholesterol content, whereas mt-membranes with lower cholesterol content are affected only at higher concentrations. Digitonin is a natural product and thus the effective concentration has to be determined by titrations for every batch. The optimum effective digitonin concentrations for complete plasma membrane permeabilization of cultured cells can be determined directly in a respirometric protocol (see: SUIT-010 O2 ce-pce D008). |
| Dihydro-orotate dehydrogenase | DhoDH | Dihydro-orotate dehydrogenase is an electron transfer complex of the inner mitochondrial membrane, converting dihydro-orotate (Dho) into orotate, and linking electron transfer through the Q-junction to pyrimidine synthesis and thus to the control of biogenesis. |
| Dihydroethidium | DHE | Dihydroethidium (also called hydroethidine) is a cell permeant fluorescent probe used to analyse superoxide presence. It is a reduced form of ethidium that presents blue fluorescence, and after oxidation by superoxide becomes able to intercalate DNA and emits red fluorescence (excitation wavelength ~518–535 nm, emission ~605–610 nm). It has been used to detect superoxide by HPLC and by fluorescence microscopy. |
| Dilution effect | Dilution of the concentration of a compound or sample in the experimental chamber by a titration of another solution into the chamber. | |
| Dimension | Dimensions are defined in the SI {Quote}: Physical quantities can be organized in a system of dimensions, where the system used is decided by convention. Each of the seven base quantities used in the SI is regarded as having its own dimension. .. All other quantities, with the exception of counts, are derived quantities, which may be written in terms of base quantities according to the equations of physics. The dimensions of the derived quantities are written as products of powers of the dimensions of the base quantities using the equations that relate the derived quantities to the base quantities. There are quantities Q for which the defining equation is such that all of the dimensional exponents in the equation for the dimension of Q are zero. This is true in particular for any quantity that is defined as the ratio of two quantities of the same kind. .. There are also some quantities that cannot be described in terms of the seven base quantities of the SI, but have the nature of a count. Examples are a number of molecules, a number of cellular or biomolecular entities (for example copies of a particular nucleic acid sequence), or degeneracy in quantum mechanics. Counting quantities are also quantities with the associated unit one. {end of Quote: p 136, Bureau International des Poids et Mesures 2019 The International System of Units (SI)} | |
| Dimethyl sulfoxide | DMSO | Dimethyl sulfoxide is a polar aprotic solvent that dissolves both polar and nonpolar compounds and is miscible in a wide range of organic solvents as well as water. DMSO may also be used as a cryoprotectant, added to cell media to reduce ice formation and thereby prevent cell death during the freezing process. |
| Dinitrochlorobenzene | DNCB | Dinitrochlorobenzene (1-chloro-2,4-dinitrobenzene) (DNCB) is a glutathione (GSH) inhibitor. |
| Dinitrophenole | DNP | 2,4-dinitrophenole (C6H4N2O5; M = 184.11 g·mol-1) is a protonophore acting as an uncoupler of oxidative phosphorylation. |
| Directive | A directive is a legal act of the European Union, which requires member states to achieve a particular result without dictating the means of achieving that result. | |
| Directory of Open Access Journals | DOAJ | The Directory of Open Access Journals is a free online directory that indexes and provides access to open access peer-reviewed journals. |
| Discontinuous system | In a discontinuous system, gradients in continuous systems across the length, l, of the diffusion path [m], are replaced by differences across compartmental boundaries of zero thickness, and the local concentration is replaced by the free activity, α [mol·dm-3]. The length of the diffusion path may not be constant along all diffusion pathways, spacial direction varies (e.g., in a spherical particle surrounded by a semipermeable membrane), and information on the diffusion paths may even be not known in a discontinuous system. In this case (e.g., in most treatments of the protonmotive force) the diffusion path is moved from the (ergodynamic) isomorphic force term to the (kinetic) mobility term. The synonym of a discontinuous system is compartmental or discretized system. In the first part of the definition of discontinuous systems, three compartments are considered: (1) the source compartment A, (2) the sink compartment B, and (3) the internal barrier compartment with thickness l. In a two-compartmental description, a system boundary is defined of zero thickness, such that the barrier comparment (e.g., a semipermeable membrane) is either part of the system (internal) or part of the environment (external). Similarly, the intermediary steps in a chemical reaction may be explicitely considered in an ergodnamic multi-comparment system; alternatively, the kinetic analysis of all intermediary steps may be collectively considered in the catalytic reaction mobility, reducing the measurement to a two-compartmental analysis of the substrate and product compartments. | |
| Dispersion devices | A dispersion device diffracts light at different angles according to its wavelength. Traditionally, prisms and diffraction gratings are used, the latter now being the most commonly used device in a spectrofluorometer or spectrophotometer. | |
| Display DatLab help | Display DatLab help
In this section, we present some issues that could happen during your data analysis related to the graphs display and how to fix them quickly. Case in which an issue might occur:
In the event of a frozen display of the graphs, try the alternatives below:
| |
| Display Power-O2k | The Power-O2k number, which is set in the pull-down menu Oroboros O2k \ O2k configuration, is shown in the active graph. To show it in graphs copied to clipboard, the option "Show Oroboros icon in clipboard files" must be enabled in the Graph-menu Graph options - DatLab. | |
| Display numerical value | If Display numerical value the current numerical values are displayed in the graph for the active plots on the Y1 axis and Y2 axis (during data acquisition only). | |
| Dithionite | Dit Dit - (The abbreviation 'Dith' has been used previously and is stepwise replaced by Dit.) | The sodium salt of Dithionite Na2S2O4 (Dit) is the 'zero oxygen solution powder' used for calibration of oxygen sensors at zero oxygen concentration, or for stepwise reduction of oxygen concentrations in instrumental O2 background tests. It is not recommended to use dithionite in experiments with biological samples or several multisensor approaches, for these see Setting the oxygen concentration. |
| Drift | The most common cause of drift is variation in the intensity of the light source. The effect of this can be minimised by carrying out a balance at frequent intervals. | |
| Dual wavelength analysis | If a sample contains a number of absorbing substances, it is sometimes possible to select discrete pairs of wavelengths at which the change in absorbance of a particular substance (due to oxidation or reduction, for example) is largely independent of changes in the absorbance of other substances present. Dual wavelength analysis can be carried out for cytochrome c by subtracting the absorbance at 540 nm from that at 550nm in order to give a measure of the degree of reduction. Similarly, by subtracting the absorbance at 465 nm from that at 444 nm, an indicator of the redox state of cytochrome aa3 can be obtained. | |
| Duroquinol | DQ | ET-pathway level 2 is supported by duroquinol DQ feeding electrons into Complex III (CIII) with further electron transfer to CIV and oxygen. Upstream pathways are inhibited by rotenone and malonic acid in the absence of other substrates linked to ET-pathways with entry into the Q-junction. |
| Dyscoupled respiration | Dyscoupled respiration is LEAK respiration distinguished from intrinsically (physiologically) uncoupled and from extrinsic experimentally uncoupled respiration as an indication of extrinsic uncoupling (pathological, toxicological, pharmacological by agents that are not specifically applied to induce uncoupling, but are tested for their potential dyscoupling effect). Dyscoupling indicates a mitochondrial dysfunction. In addition to intrinsic uncoupling, dyscoupling occurs under pathological and toxicological conditions. Thus a distinction is made between physiological uncoupling and pathologically defective dyscoupling in mitochondrial respiration. | |
| E | e, E | » Energy, Exergy E
» elementary charge e = 1.602 176 634∙10-19 C∙x-1 » Euler's number e ~ 2.718 281 828 459 » ET capacity E |
| E-L coupling efficiency | jE-L | |
| E-L net ET capacity | E-L | |
| E-P control efficiency | jE-P | |
| E-P excess capacity | E-P | |
| E-R control efficiency | jE-R | |
| E-R reserve capacity | E-R | |
| ET capacity | E | |
| ET-pathway competent state | Electron transfer pathway competent state, see Electron-transfer-pathway state. | |
| ET-pathway substrate types | n.a. | See Electron-transfer-pathway state |
| EUROMIT | EUROMIT is a group based in Europe for organizing International Meetings on Mitochondrial Pathology. | |
| Ectotherms | Ectotherms are organisms whose body temperatures conform to the thermal environment. In many cases, therefore, ectotherms are poicilothermic. | |
| Editorial board participation | Editorial board participation is a topic addressed in COPE core practices for research. | |
| Elamipretide | Bendavia | Bendavia (Elamipretide) was developed as a mitochondria-targeted drug against degenerative diseases, including cardiac ischemia-reperfusion injury. Clinical trials showed variable results. It is a cationic tetrapeptide which readily passes cell membranes, associates with cardiolipin in the mitochondrial inner membrane. Supercomplex-associated CIV activity significantly improved in response to elamipretide treatment in the failing human heart. |
| Elasticity | ε | According to David Fell, "Elasticities are properties of individual enzymes and not the metabolic system. The elasticity of an enzyme to a metabolite is related to the slope of the curve of the enzyme's rate plotted against metabolite concentration, taken at the metabolite concentrations found in the pathway in the metabolic state of interest. It can be obtained directly as the slope of the logarithm of the rate plotted against the logarithm of the metabolic concentration. The elasticity will change at each point of the curve (s,v) and must be calculated for the specific concentration of the metabolite (s) that will give a specific rate (r) of the enzyme activity" (See Figure).
 |
| Electric current | Iel [A = C·s-1]; [mol·s-1]; [x·s-1] | Current or electric flow Iel is the advancement of charge per unit of time, expressed in the SI base unit ampere [C·s-1 = A]. Electrons or ions are the current-carrying motive entities of electric flow. Electrons e- are negatively charged subatomic particles carrying 'negative electricity' with a mass that is about 1/1700 of the smallest particle — the proton — carrying 'positive electricity' (Thompson 1906). Correspondingly the velocity of electrons is much higher than that of protons or any other (larger) ion. Current is the velocity v of paticles times the number of motive charges. Therefore, electron current Ie- is of a different nature from electric current Ielχ carried by all species i of ions Xi (cations and anions) summarized as χ = Σ(zi·Xi). Whereas Ie- is the net translocation of electrons moving forwards and backwards, Ielχ is the net translocation of charges carried by different cations and anions. In contrast, ion current IelX of a specific ion X is the partial translocation of charges carried by net translocation of ion X only. If cation current IelX+ is antagonized entirely by counterion current IelY- as the process of antiport, then the electric current Ielχ is zero. The (net) electric current in a compartmental system is driven by the electric force ΔelFp+ or electric potential difference ΔΨp+, whereas a compensated ion/counterion antiport current is insensitive to the electric potential difference. |
| Electric current density | j [C·m-2] | Electric current density is current divided by area, j=I·A-1 [C·m-2]. Compare: density. |
| Electrochemical constant | f [J·C-1·K-1] | The electrochemical constant f has the SI unit for energy per charge per temperature [J·C-1·K-1].
f = k·e-1, the Boltzmann constant k divided by the elementary charge e. f = R·F-1, the gas constant R divided by the Faraday constant F. |
| Electrolyte\Reference-Electrode | Electrolyte\Reference-Electrode for Reference-Electrode\2.4 mm | |
| Electron flow | Ie | Electron flow through the mitochondrial Electron transfer pathway (ET-pahway) is the scalar component of chemical reactions in oxidative phosphorylation (OXPHOS). Electron flow is most conveniently measured as oxygen consumption (oxygraphic measurement of oxygen flow), with four electrons being taken up when oxygen (O2) is reduced to water. |
| Electron leak | Electrons that escape the electron transfer pathway without completing the reduction of oxygen to water at cytochrome c oxidase, causing the production of ROS. The rate of electron leak depends on the topology of the complex, the redox state of the moiety responsible of electron leakiness and usually on the protonmotive force (Δp). In some cases, the Δp dependance relies more on the ∆pH component than in the ∆Ψ. | |
| Electron transfer pathway | ET pathway | In the mitochondrial electron transfer pathway (ET pathway) electrons are transferred from externally supplied reduced fuel substrates to oxygen. Based on this experimentally oriented definition (see ET capacity), the ET pathway consists of (1) the membrane-bound ET pathway with respiratory complexes located in the inner mt-membrane, (2) TCA cycle and other mt-matrix dehydrogenases generating NADH and succinate, and (3) the carriers involved in metabolite transport across the mt-membranes. » MiPNet article |
| Electron-transfer-pathway state | ET-pathway state |
Electron-transfer-pathway states are obtained in mitochondrial preparations (isolated mitochondria, permeabilized cells, permeabilized tissues, tissue homogenate) by depletion of endogenous substrates and addition to the mitochondrial respiration medium of fuel substrates (CHNO) activating specific mitochondrial pathways, and possibly inhibitors of specific pathways. Mitochondrial electron-transfer-pathway states have to be defined complementary to mitochondrial coupling-control states. Coupling-control states require ET-pathway competent states, including oxygen supply. Categories of SUIT protocols are defined according to mitochondrial ET-pathway states. » MiPNet article |
| Electron-transferring flavoprotein Complex | CETF | Electron-transferring flavoprotein Complex (CETF) is a respiratory Complex localized at the matrix face of the inner mitochondrial membrane, supplies electrons to Q, and is thus an enzyme Complex of the mitochondrial Electron transfer pathway (ET-pathway). CETF links the ß-oxidation cycle with the membrane-bound electron transfer system in fatty acid oxidation (FAO). |
| Electronic-TIP2k Upgrading\O2k-Main Unit Series A-D | Electronic-TIP2k Upgrading\O2k-Main Unit Series A-D - Former Product : not required for O2k-Core, the O2k-Main Unit has to be returned to the OROBOROS workshop. | |
| Electronic-TIP2k Upgrading\O2k-Main Unit Series E | Electronic-TIP2k Upgrading\O2k-Main Unit Series E - Former Series : not required for O2k-Core, free of charge for Series E in conjunction with the purchase of the TIP2k-Module, the O2k-Main Unit has to be returned to the OROBOROS workshop. | |
| Elementary charge | e [C·x-1] | The elementary charge or proton charge e has the SI unit coulomb [C], but more strictly coulomb per elementary unit [C·x-1]. -e is the charge per electron. Elementary charge e is the charge per elementary entity H+ with SI unit [C] but canonical SI unit [C·x-1]. When the charge Qel [C] of a number Ne [x] of electrons e is divided by the count Ne, then the particle charge QNX (QNX) charge per elementary entity is obtained, -e = Qel/Ne [C·x-1]. e is also used as an atomic unit. |
| Elementary entity | UX [x] |
An elementary entity is an entity of type X, distinguished as a single unit of countable objects (X = molecules, cells, organisms, particles, parties, items) or events (X = beats, collisions, emissions, decays, celestial cycles, instances, occurrences, parties). "An elementary entity may be an atom, a molecule, an ion, an electron, any other particle or specified group of particles" (Bureau International des Poids et Mesures 2019). An elementary entity, therefore, needs to be distinguished from non-countable entities and the general class of entities X. This distinction is emphasized by the term 'elementary' (synonymous with 'elementary entity') with symbol UX and elementary unit [x]. If an object is defined as an assembly of particles (a party of two, a molecule as the assembly of a stoichiometric number of atoms), then the elementary is the assembly but not the assembled particle. A number of defined elementaries UX is a count, NX = N·UX [x], where N is a number, and as such N is dimensionless, and N is a number (stop) and is not 'a number of ..'. Elementaries are added as items to a count. The elementary UX has the dimension U of the count NX. The elementary UX has the same unit [x] as the count NX, or more accurately it gives the count the defining 'counting-unit', which is the 'elementary unit' [x]. From the definition of count as the number (N) of elementaries (U) of entity type X, it follows that count divided by elementary is a pure number, N = NX·UX-1. The unit x of a count can neither be the entity X nor a number. The elementary of type X defines the identity X of the elementary UX with the unit 'elementary unit' with symbol [x]. Since a count NX is the number of elementary entities, the elementary UX is not a count (UX is not identical with N·UX). |
| Elementary unit | x | The elementary unit [x] is the unit of a count NX [x]. The International System of Units defines the unit of a count as 1. Then the Number 1 is the Unit of the Count of Entities — NUCE. This causes a number of formal inconsistencies which are resolved by introducing the elementary unit [x] as the abstracted unit of Euclid’s unit, which is an elementary entity UX [x], and as the unit of Euclid’s number, which is a count NX [x]. |
| Enable DL-Protocol editing | Enable DL-Protocol editing is a novel function of DatLab 7.4 offering a new feature in DL-Protocols: flexibility. Fixed sequences of events and marks can be changed (Skip/Added) in a SUIT protocol by the user. Moreover, the text, instructions, concentrations and titration volumes of injections in a specific DL-Protocol can be edited and saved as user-specific DL-Protocol [File]\Export\DL-Protocol User (*.DLPU). To enable it, under the 'Protocols' tab in the menu, select the option 'Enable DL-Protocol editing', and then select the plot in which the marks will be set (e.g., O2 flux per V). Select the 'Overview' window, where you will be able to edit events and marks names, definition/state, final concentration and titration volumes, as well as select a mark as 'multi' for multiple titration steps, skip a mark, or add a new event or mark. After saving, export a DL-Protocol User (DLPU) and load it before running the next experiments. If users of DatLab versions older than DatLab 7.4 wish to alter the nature of the chemicals used or the sequence of injections, we ask them to contact the O2k-Technical Support.
For more information:  Export DL-Protocol User (*.DLPU) Export DL-Protocol User (*.DLPU) | |
| Endergonic | Endergonic transformations or processes can proceed in the forward direction only by coupling to an exergonic process with a driving force more negative than the positive force of the endergonic process. The backward direction of an endergonic process is exergonic. The distinction between endergonic and endothermic processes is at the heart of ergodynamics, emphasising the concept of exergy changes, linked to the performance of work, in contrast to enthalpy changes, linked to heat or thermal processes, the latter expression being terminologically linked to thermodynamics. | |
| Endothermic | An energy transformation is endothermic if the enthalpy change of a closed system is positive when the process takes place in the forward direction and heat is absorbed from the environment under isothermal conditions (∆eQ > 0) without performance of work (∆eW = 0). The same energy transformation is exothermic if it proceeds in the backward direction. Exothermic and endothermic transformations can proceed spontaneously without coupling only, if they are exergonic. | |
| Endothermy | Endothermy is the constant regulation of body temperature by metabolic heat production and control of heat exchange with the environment. | |
| Energy | E; various [J] | Heat and work are forms of energy [1 cal = 4.184 J]. Energy [J] is a fundamental term that is used in physics and physical chemistry with various meanings [1]. These meanings become explicit in the following equations relating to systems at constant volume (dV = 0) or constant gas pressure (dp = 0). Energy is exchanged between a system and the environment across the system boundaries in the form of heat, deQ, total or available work, detW (or detW), and matter, dmatU (or dmatH) [2],
dU = (deQ + detW) + dmatU ; dV = 0 [Eq. 1a] dH = (deQ + deW) + dmatH ; dp = 0 [Eq. 1b] Whereas dU (or dH) describe the internal-energy change (or enthalpy change) of the system, heat and work are external energy changes (subscript e; et: external total; e: external excluding pressure-volume work), and dmatU (or dmatH) are the exchange of matter expressed in internal-energy (or enthaply) equivalents. In closed systems, dmatU = 0 (dmatH = 0). The energy balance equation [Eq. 1] is a form of the First Law of Thermodynamics, which is the law of conservation of internal-energy, stating that energy cannot be generated or destroyed: energy can only be transformed into different forms of work and heat, and transferred in the form of matter. Notably, the term energy is general and vague, since energy may be associated with either the first or second law of thermodynamics. Work is a form of energy exchange [Eq. 1], but can be seen as exergy exchange in conjunction with deG = deW in a closed system [Eq. 3b]. An equally famous energy balance equation considers energy changes of the system only, in the most simple form for isothermal systems (dT = 0): dU = dA + T∙dS = dU + dB [Eq. 2a] dH = dG + T∙dS = dG + dB [Eq. 2b] The internal-energy change, dU (enthalpy change, dH) is the sum of free energy change (Helmholtz energy, dA; or Gibbs energy = exergy change, dG) and bound energy change (bound energy, dB = T∙dS). The bound energy is that part of the energy change that is always bound to an exchange of heat. A third energy balance equation accounts for changes of the system in terms of irreversible internal processes (i) occuring within the system boundaries, and reversible external processes (e) of transfer across the system boundaries (at constant gas pressure), dH = diH + deH [Eq. 3a] dG = diG + deG [Eq. 3b] The energy conservation law of thermodynamics (first law) can be formulated as diH = 0 (at constant gas pressure), whereas the generally negative sign of the dissipated energy, diG ≡ diD ≤ 0, is a formulation of the second law of thermodynamics. Insertion into Eq. 3 yields, dH = deH [Eq. 4a] dG = diD + deW + dmatG [Eq. 4b]When talking about energy transformations, the term energy is used in a general sense without specification of these various forms of energy. |
| Energy charge | AEC | The energy charge of the adenylate system or adenylate energy charge (AEC) has been defined by Atkinson and Walton (1967) as (ATP + ½ ADP)/(AMP + ADP + ATP). Wheather the AEC is a fundamental metabolic control parameter remains a controversial topic. |
| Energy metabolism | Core energy metabolism is the integrated biochemical process supplying the cell with ATP, utilizing ATP for various forms of work including biogenesis, maintaining ion and redox balance, and in specific organisms or tissues dissipating heat for temperature regulation. | |
| Energy saving in research | Energy saving in research must rank as a priority of social responsibility — ever since the Club of Rome published 50 years ago the seminal book on The limits to growth (1972) [1], and more so today in face of the global threat of climate change and the russian war in aggression against Ukraine.
Energy saving in research does not and must not clash with quality in research. Application of high-quality and predefined experimental protocols combined with evaluation of repeatability and reproducibility represents primary strategies for energy saving in research. Publication of irreproducible results — adding to the reproducibility crisis — is the most wasteful aspect of research in terms of resources including energy (more properly: exergy). Paywall journalism is wasteful in terms of financial resources. Dramatically increasing numbers of scientific publications is a pathway towards waste of energy [2]. Besides large-scale strategies on e(n)xergy saving in research — quality versus quantity —, everybody's everyday contributions to energy saving count: to cut greenhouse gas emissions, save biological and geological diversity, and improve equality across societies, gender, continents, and countries. Do scientists take responsibility for energy saving? Or does biomedical research merely find excuses? Scientific institutions in academia and industry must implement energy saving strategies to reduce waste according to the European Union's Energy efficiency directive, and to consume less energy (exergy) by using it more efficiently (Energy efficiency targets). Possible — important but much neglected — contributions include:
| |
| Enthalpy | H [J] | Enthalpy, H [J], can under conditions of constant gas pressure neither be destroyed nor created (first law of thermodynamics: diH/dt = 0). The distinction between enthalpy and internal-energy of a system is due to external pressure-volume work carried out reversibly at constant gas pressure. The enthalpy change of the system, dH, at constant pressure, is the internal-energy change, dU, minus reversible pressure-volume work,
dH = dU - dVW Pressure-volume work, dVW, at constant pressure, is the gas pressure, p [Pa = J·m-3], times change of volume, dV [m3], dVW = -p·dV [J] The available work, deW, is distinguished from external total work, detW, [1] deW = detW - dVW The change of enthalpy of a system is due to internal and external changes, dH = diH + deH Since diH = 0 (first law of thermodynamics), the dH is balanced by exchange of heat, work, and matter, dH = (deQ + deW) + dmatH ; dp = 0 The exchange of matter is expressed in enthalpy equivalents with respect to a reference state (formation, f, or combustion, c). The value of dH in an open system, therefore, depends on the arbitrary choice of the reference state. In contrast, the terms in parentheses are the sum of all (total, t) partial energy transformations, dtH = (deQ + deW) A partial enthalpy change of transformation, dtrH, is distinguished from the total enthalpy change of all transformations, dtH, and from the enthalpy change of the system, dH. In a closed system, dH = dtH. The enthalpy change of transformation is the sum of the Gibbs energy (free energy) change of transformation, dtrG, and the bound energy change of transformation at constant temperature and pressure, dtrB = T·dS, dtrH = dtrG + dtrB |
| Entity | X | An entity of type X is something that can measured as an extensive quantity or counted as an elementary entity. The term entity with symbol X, therefore, has a general meaning, including but not limited to elementary entities UX. The distinction can be emphasized by using the term entity-type X, to avoid confusion of an entity X with the more restricted definition of elementary entity UX as a single countable object or event. |
| Equality | = | Physicochemical equality (symbol =) indicates in an equation not only numerical equivalence (symbol ≡), but an identity of the full meaning. |
| Equivalence | ≡ | Numerical equivalence (symbol ≡) indicates that two quantities are numerically equal, even if the full meaning may be different. For instance: 1 ≡ 1·1 and 1 ≡ 1/1. In contrast to ≡, the symbol = indicates physicochemical equality. |
| Ergodynamic efficiency | ε | The ergodynamic efficiency, ε (compare thermodynamic efficiency), is a power ratio between the output power and the (negative) input power of an energetically coupled process. Since power [W] is the product of a flow and the conjugated thermodynamic force, the ergodynamic efficiency is the product of an output/input flow ratio and the corresponding force ratio. The efficiency is 0.0 in a fully uncoupled system (zero output flow) or at level flow (zero output force). The maximum efficiency of 1.0 can be reached only in a fully (mechanistically) coupled system at the limit of zero flow at ergodynamic equilibrium. The ergodynamic efficiency of coupling between ATP production (DT phosphorylation) and oxygen consumption is the flux ratio of DT phosphorylation flux and oxygen flux (P»/O2 ratio) multiplied by the corresponding force ratio. Compare with the OXPHOS-coupling efficiency. |
| Ergodynamics | The mission of ergodynamics is the revelation of relations of general validity. "Thermodynamics deals with relationships between properties of systems at equilibrium and with differences in properties between various equilibrium states. It has nothing to do with time. Even so, it is one of the most powerful tools of physical chemistry" [1]. Ergodynamics is the theory of exergy changes (from the Greek word 'erg' which means work). Ergodynamics includes the fundamental aspects of thermodynamics ('heat') and the thermodynamics of irreversible processes (TIP; nonequilibrium thermodynamics), and thus links thermodynamics to kinetics. In its most general scope, ergodynamics is the science of energy transformations. Classical thermodynamics includes open systems, yet as a main focus it describes closed systems. This is reflected in a nomenclature that is not easily applicable to the more general case of open systems [2]. At present, IUPAC recommendations [3] fall short of providing adequate guidelines for describing energy transformations in open systems. | |
| Ethanol | ethanol abs. |
Ethanol or ethyl alcohol, C2H6O or EtOH, is widely used in the laboratory, particularly as a solvent and cleaning agent. There are different grades of high purity ethanol. Up to a purity of 95.6 % ethanol can be separated from water by destillation. Higher concentrations than 95% require usage of additives that disrupt the azeotrope composition and allow further distillation. Ethanol is qualified as "absolute" if it contains no more than one percent water. Whenever 'ethanol abs.' is mentioned without further specification in published protocols, it refers to ≥ 99 % ethanol a.r. (analytical reagent grade).
|
| Ethics on publishing | Ethics on publishing follow COPE's guidelines (or equivalent). A journal's policy on publishing ethics should be clearly visible on its website, and should refer to: (1) Journal policies on authorship and contributorship; (2) How the journal will handle complaints and appeals; (3) Journal policies on conflicts of interest / competing interests; (4) Journal policies on data sharing and reproducibility; (5) Journal's policy on ethical oversight; (6) Journal's policy on intellectual property; and (7) Journal's options for post-publication discussions and corrections. | |
| Ethylene glycol tetraacetic acid | EGTA | Ethylene glycol tetraacetic acid (EGTA) is a chelator for heavy metals, with high affinity for Ca2+ but low affinity for Mg2+. Sigma E 4378. |
| Etomoxir | Eto | Etomoxir (Eto; 2[6(4-chlorophenoxy)hexyl]oxirane-2-carboxylate) is an irreversible inhibitor of carnitine palmitoyltransferase I (CPT-I) on the outer face of the mitochondrial inner membrane. Eto inhibits fatty acid oxidation by blocking the formation of acyl carnitines from long-chain fatty acids which require the carnitine shuttle for transport into mitochondria. In contrast to long-chain fatty acids, the transport of short- and medium-chain fatty acids is carnitine-independent. |
| European Bioenergetics Conference |  EBEC is a group based in Europe that organizes the European Bioenergetics Conference.
EBEC is a group based in Europe that organizes the European Bioenergetics Conference. | |
| Euthanyl/Pentobarbitol | Euthanyl | I am often asked by reviewers to discuss the effects of pentobarbitol euthansia on mithochondrial function. Takaki 1997 JJP: This paper has been helpful in this discussion. (edit by Staples JF) |
| Events - DatLab | F4 | An event in DatLab is a defined point in time, labelled by a name (1 to 10 characters). An event applies to all plots of the selected O2k-Chamber. The event is shown by a vertical line in the graph and the label of the event is shown on the top (DatLab 6 and lower: on the bottom). The default name is the sequential number of the event. It is recommended to edit event labels with a minimum number of characters, and to explain the abbreviation in the 'Definition' box. The final concentration and titration volume can be entered into the corresponding boxes, if the event relates to the titration of a substance. A short comment can be entered to describe the event in detail.
Set events - Manual events are entered (real-time, connected to the O2k) by pressing [F4] at the time of the event (e.g. to indicate a manual titration into the chamber). An event belongs either to chamber A, chamber B, or both. Instrumental events are added automatically, e.g. when the stirrer (A or B) or illumination (both chambers) is switched on or off. After setting a new event the Edit event window pops up. Pressing F4 defines the time point of the event. Full attention can then be paid to the experiment. Edit the event later, as it is possible to insert an event at any chosen moment of the plotted record of the experiment by placing the cursor anywhere in the graph at the selected time point by pressing Ctrl and clicking the left mouse button. Edit event - Left click on the name of an existing event to open the Edit event window to edit or Delete event. In events obtained from a selected protocol, the entire sequence of consecutive events is defined with event names, definitions, concentrations and titration volumes. Name - Enter an event name of 1 to 10 characters. Short names (e.g. O instead of Open) are recommended. Comment - Further information can be entered into the text field. Select O2k-chamber A, B or both. The Event will be shown on plots for both or one selected chamber. »Protocol events |
| Examination | An examination is a set of operations having the object of determining the value or characteristics of a property. In some disciplines (e.g. microbiology) an examination is the total activity of a number of tests, observations or measurements. | |
| Exclusion criteria | The Exclusion criteria include factors or characteristics that make the recruited population ineligible for the outcome parameter. With the Inclusion criteria, this factor must be a cofounder for the outcome parameter | |
| Exergonic | Exergonic transformations or processes can spontaneously proceed in the forward direction, entailing the irreversible loss of the potential to performe work (erg) with the implication of a positive internal entropy production. Ergodynamic equilibrium is obtained when an exergonic (partial) process is compensated by a coupled endergonic (partial) process, such that the Gibbs energy change of the total transformation is zero. Final thermodynamic equilibrium is reached when all exergonic processes are exhausted and all forces are zero. The backward direction of an exergonic process is endergonic. The distinction between exergonic and exothermic processes is at the heart of ergodynamics, emphasising the concept of exergy changes, linked to the performance of work, in contrast to enthalpy changes, linked to heat or thermal processes, the latter expression being terminologically linked to thermodynamics. | |
| Exergy | E; various [J] | Exergy includes external and internal work. Exergy as the external work is defined in the First Law of thermodynamics as a specific form of energy. Exergy as the dissipated Gibbs or Helmholtz energy is the irreversibly dissipated (internal) loss of the potential of performing work as defined in the Second Law of Thermodynamics.
Changes of exergy dG plus bound energy yield the enthalpy change: dH = dG + T∙dS = dG + dB |
| Exothermic | An energy transformation is exothermic if the enthalpy change of a closed system is negative when the process takes place in the forward direction and heat is lost to the environment under isothermal conditions (∆eQ < 0) without performance of work (∆eW = 0). The same energy transformation is endothermic if it proceeds in the backward direction. Exothermic and endothermic transformations can proceed spontaneously without coupling only, if they are exergonic. | |
| Experiment | A number of replica, N, of experiments on one sample type is designed to obtain statistical information about the involved population(s) and to test hypotheses about a population and about differences between populations, when experiments are carried out on different sample types. An experiment may involve various assays, e.g., a respirometric assay and an assay for protein determination. | |
| Experimental code | F3 | An experimental code can be entered in the Sample window, containing up to 10 digits. |
| Experimental log - DatLab | Ctrl+F3 | Experimental log provides an automatically generated experimental protocol with detailed information about the O2k settings and calibrations, the Sample information and various Events. Time-dependent information can be viewed for a single chamber or both chambers. The filter can be selected for viewing minimum information, intermittent by default, or all information. The experimental log can be viewed and saved as a PDF file by clicking on [Preview]. |
| Export DL-Protocol User (*.DLPU) | it is a function of DatLab (available from version 7.4 onwards) that enables the export of user specific protocols (DL-Protocol User) to the SUIT protocol folder from which they can be uploaded for subsequent measurements. | |
| Export as CSV - DatLab | Ctrl-E | Export as CSV (*.csv) exports plots and events to a text file for further use in Excel and other programs. |
| Extended abstracts | In the context of MiPevents, extended abstracts are accepted for preprint publication in MitoFit Preprints upon evaluation by the MitoFit Preprints Scientific Advisory Board. Publishing extended abstracts with MitoFit Preprints does not preclude later full journal publication, but will make your work fully citable, by assigning each manuscript a unique DOI number, and facilitate discovery and feedback. | |
| Extensive quantity | Extensive quantities pertain to a total system, e.g. oxygen flow. An extensive quantity increases proportional with system size. The magnitude of an extensive quantity is completely additive for non-interacting subsystems, such as mass or flow expressed per defined system. The magnitude of these quantities depends on the extent or size of the system (Cohen et al 2008). | |
| External flow | Ie [MU·s-1] | External flows across the system boundaries are formally reversible. Their irreversible facet is accounted for internally as transformations in a heterogenous system (internal flows, Ii). |
| Extinction | Extinction is a synonym for absorbance. | |
| Extinction coefficient | ε | The extinction coefficient (ε) of a substance is the absorbance of a 1 µmolar concentration over a 1 cm pathlength and is wavelength-dependent. |
| Extrinsic fluorophores | Extrinsic fluorophores are molecules labelled with a fluorescent dye (as opposed to intrinsic fluorescence or autofluorescence of molecules which does not require such labelling). They are available for a wide range of parameters including ROS (H2O2, Amplex red) (HOO-, MitoSOX) , mitochondrial membrane potential (Safranin, JC1, TMRM, Rhodamine 123), Ca2+ (Fura2, Indo 1, Calcium Green), pH (Fluorescein, HPTS, SNAFL-1), Mg2+ (Magnesium Green) and redox state (roGFP). | |
| Extroduction | The term extroduction is ambiguous and needs introduction. An external extroduction aims at providing a specific exit that opens the door to the parent article. Once you popped up into the article box, there are various internal extroductions to push down by following hyperlinks to references, keywords, supplementary material, and to the external extroduction. Once you have pushed one level down, there may be hyperlinks to push down further (Hofstadter 1979). One needs to keep track of the links in a nested network of open tabs, to pop up all the way back for returning to the initial reference level. | |
| F-junction | The F-junction is a junction for convergent electron flow in the electron transfer pathway (ET-pathway) from fatty acids through fatty acyl CoA dehydrogenase (reduced form FADH2) to electron transferring flavoprotein (CETF), and further transfer through the Q-junction to Complex III (CIII). The concept of the F-junction and N-junction provides a basis for defining categories of SUIT protocols. Fatty acid oxidation, in the F-pathway control state, not only depends on electron transfer through the F-junction (which is typically rate-limiting) but simultaneously generates NADH and thus depends on N-junction throughput. Hence FAO can be inhibited completely by inhibition of Complex I (CI). In addition and independent of this source of NADH, the N-junction substrate malate is required as a co-substrate for FAO in mt-preparations, since accumulation of AcetylCoA inhibits FAO in the absence of malate. Malate is oxidized in a reaction catalyzed by malate dehydrogenase to oxaloacetate (yielding NADH), which then stimulates the entry of AcetylCoA into the TCA cycle catalyzed by citrate synthase. | |
| F1000Research | F1000Research is an Open Research publishing platform for life scientists, offering immediate publication of articles and other research outputs without editorial bias. All articles benefit from transparent peer review and the inclusion of all source data. It is thus not a preprint server, but posters and slides can be published without author fees. Published posters and slides receive a DOI (digital object identifier) and become citable after a very basic check by our in-house editors. | |
| FADH2 | FADH2 | FADH2 and FAD: see Flavin adenine dinucleotide. |
| FCCP | FCCP | FCCP (Carbonyl cyanide p-trifluoro-methoxyphenyl hydrazone, C10H5F3N4O) is a protonophore or uncoupler: added at uncoupler concentration Uc; c is the optimum uncoupler concentration in titrations to obtain maximum mitochondrial respiration in the noncoupled state of ET capacity. |
| FN | FN | FN is induced in mt-preparations by addition of NADH-generating substrates (N-pathway control state, or CI-linked pathway control) in combination with one or several fatty acids, which are supplied to feed electrons into the F-junction through fatty acyl CoA dehydrogenase (reduced form FADH2), to electron transferring flavoprotein (CETF), and further through the Q-junction to Complex III (CIII). FAO not only depends on electron transfer through the F-junction (which is typically rate-limiting), but simultaneously generates FADH2 and NADH and thus depends on N-junction throughput. Hence FAO can be inhibited completely by inhibition of Complex I (CI). This physiological substrate combination is required for partial reconstitution of TCA cycle function and convergent electron-input into the Q-junction, to compensate for metabolite depletion into the incubation medium. FS in combination exerts an additive effect of convergent electron flow in most types of mitochondria. |
| FNS | FNS | FNS is induced in mt-preparations by addition of NADH-generating substrates (N-pathway control state, or CI-linked pathway control) in combination with succinate (S-pathway control state; S- or CII-linked) and one or several fatty acids, which are supplied to feed electrons into the F-junction through fatty acyl CoA dehydrogenase (reduced form FADH2), to electron transferring flavoprotein (CETF), and further through the Q-junction to Complex III (CIII). FAO not only depends on electron transfer through the F-junction (which is typically rate-limiting), but simultaneously generates FADH2 and NADH and thus depends on N-junction throughput. Hence FAO can be inhibited completely by inhibition of Complex I (CI). This physiological substrate combination is required for partial reconstitution of TCA cycle function and convergent electron-input into the Q-junction, to compensate for metabolite depletion into the incubation medium. FNS in combination exerts an additive effect of convergent electron flow in most types of mitochondria. |
| FNSGp | FNSGp |
MitoPathway control state: FNSGp
SUIT protocol: SUIT-002 This substrate combination supports convergent electron flow to the Q-junction. |
| Faraday constant | F [C/mol] | The Faraday constant F links the electric charge [C] to amount [mol], and thus relates the electrical format e [C] to the molar format n [mol]. The Farady constant, F = e·NA = 96 485.33 C/mol, is the product of elementary charge, e = 1.602176634∙10-19 C/x, and the Avogadro constant, NA = 6.02214076∙1023 x/mol. The dimensionless unit [x] is not explicitely considered by IUPAC. |
| Fatty acid | FA | Fatty acids are carboxylic acids with a carbon aliphatic chain. The fatty acids can be divided by the length of this chain, being considered as short-chain (1–6 carbons), medium-chain (7–12 carbons) and long-chain and very long-chain fatty acids (>12 carbons).
Long-chain fatty acids must be bound to carnitine to enter the mitochondrial matrix, in a reaction that can be catalysed by carnitine acyltransferase. For this reason, long-chain fatty acids, such as palmitate (16 carbons) is frequently supplied to mt-preparations in the activated form of palmitoylcarnitine. Fatty acids with shorter chains, as octanoate (8 carbons) may enter the mitochondrial matrix, however, in HRR they are more frequently supplied also in the activated form, such as octanoylcarnitine. Once in the mitochondrial matrix, the fatty acid oxidation (FAO) occurs, generating acetyl-CoA, NADH and FADH2. In the fatty acid oxidation pathway control state electrons are fed into the F-junction involving the electron transferring flavoprotein (CETF). FAO cannot proceed without a substrate combination of fatty acids & malate, and inhibition of CI blocks FAO. Low concentration of malate, typically 0.1 mM, does not saturate the N-pathway; but saturates the F-pathway. |
| Fatty acid oxidation | FAO | Fatty acid oxidation is a multi-step process by which fatty acids are broken down in β-oxidation to generate acetyl-CoA, NADH and FADH2 for further electron transfer to CoQ. Whereas NADH is the substrate of CI, FADH2 is the substrate of electron-transferring flavoprotein complex (CETF) which is localized on the matrix face of the mtIM, and supplies electrons from FADH2 to CoQ. Before the ß-oxidation in the mitochondrial matrix, fatty acids (short-chain with 1-6, medium-chain with 7–12, long-chain with >12 carbon atoms) are activated by fatty acyl-CoA synthases (thiokinases) in the cytosol. For the mitochondrial transport of long-chain fatty acids the mtOM-enzyme carnitine palmitoyltransferase I (CPT-1; considered as a rate-limiting step in FAO) is required which generates an acyl-carnitine intermediate from acyl-CoA and carnitine. In the next step, an integral mtIM protein carnitine-acylcarnitine translocase (CACT) catalyzes the entrance of acyl-carnitines into the mitochondrial matrix in exchange for free carnitines. In the inner side of the mtIM, another enzyme carnitine palmitoyltransferase 2 (CPT-2) converts the acyl-carnitines to carnitine and acyl-CoAs, which undergo ß-oxidation in the mitochondrial matrix. Short- and medium-chain fatty acids do not require the carnitine shuttle for mitochondrial transport. Octanoate, but not palmitate, (eight- and 16-carbon saturated fatty acids) may pass the mt-membranes, but both are frequently supplied to mt-preparations in the activated form of octanoylcarnitine or palmitoylcarnitine. |
| Fatty acid oxidation pathway control state | F, FAO | In the fatty acid oxidation pathway control state (F- or FAO-pathway), one or several fatty acids are supplied to feed electrons into the F-junction through fatty acyl CoA dehydrogenase (reduced form FADH2), to electron transferring flavoprotein (CETF), and further through the Q-junction to Complex III (CIII). FAO not only depends on electron transfer through the F-junction (which is typically rate-limiting relative to the N-pathway branch), but simultaneously generates FADH2 and NADH and thus depends on N-junction throughput. Hence FAO can be inhibited completely by inhibition of Complex I (CI). In addition and independent of this source of NADH, the type N substrate malate is required at low concentration (0.1 mM) as a co-substrate for FAO in mt-preparations, since accumulation of Acetyl-CoA inhibits FAO in the absence of malate. Malate is oxidized in a reaction catalyzed by malate dehydrogenase to oxaloacetate (yielding NADH), which then stimulates the entry of Acetyl-CoA into the TCA cycle catalyzed by citrate synthase. Peroxysomal β-oxidation carries out few β-oxidation cycles, thus shortening very-long-chain fatty acids (>C20) for entry into mitochondrial β-oxidation. Oxygen consumption by peroxisomal acyl-CoA oxidase is considered as residual oxygen consumption rather than cell respiration. |
| Fermentation | Fermentation is the process of energy metabolism used to supply ATP, where redox balance is maintained with internally produced electron acceptors (such as pyruvate or fumarate), without the use of external electron acceptors (such as O2). Fermentation thus contrasts with cell respiration and is an anaerobic process, but aerobic fermentation may proceed in the presence of oxygen. | |
| File search - DatLab | Ctrl+F | File search yields a list of all files labelled by the experimental code in a selected directory . Click on the file to preview the experimental log. With File Search you can search in all folders and subfolders on your computer for DatLab files with a selected experimental code. The experimental code is entered in the DatLab file in the window "Experiment" ([F3]). When you click on a folder and press the button search, the DatLab file names will appear on the right window. Click on a DatLab file and further information (e.g. Sample information, Background information) will appear in the window below. |
| File:MitoFitPreprints and BEC manuscript template.docx | Bioenergetics Communications and MitoFit Preprints manuscript template. | |
| Filter Set AmR | Filter Set AmR: Set of filters for the determination of H2O2 production with Amplex UltraRed. These filters should be used together with Fluorescence-Sensor Green. The filter set consists of 6 LED filters (round) and 6 photodiode filters (rectangular). | |
| Filter Set MgG / CaG | Filter set MgG / CaG: Set of filters for the determination of concentraions of Mg2+ or Ca2+ with the fluorophores Magnesium green and Calcium green, respectively. These filters should be used together with Fluorescence-Sensor Blue or Smart Fluo-Sensor Blue. The filter set consists of 6 LED filters (round) and 6 photodiode filters (rectangular). | |
| Filter Set Saf | Filter set Saf: Set of filters for the (qualitative) determination of mitochondrial membrane potential with Safranin. These filters should be used together with Fluorescence-Sensor Blue or Smart Fluo-Sensor Blue. The filter set consists of 6 LED filters (round) and 6 photodiode filters (rectangular). | |
| Filter-Cap | Filter-Cap: O2k-Fluo LED2-Module (O2k-Series D to G) sensors (Fluorescence-Sensor Green and Fluorescence-Sensor Blue) and O2k-FluoRespirometer (O2k-Series H to I) sensors (Smart Fluo-Sensor Green and Smart Fluo-Sensor Blue) are equipped with a removable Filter-Cap for exchange of optical filters for the optical pathways from the LED to the sample and from the sample to the photodiode. | |
| Filters | Filters are materials that have wavelength-dependent transmission characteristics. They are can be used to select the wavelength range of the light emerging from a light source, or the range entering the detector, having passed through the sample. In particular they are used in fluorometry to exclude wavelengths greater than the excitation wavelength from reaching the sample, preventing absorption interfering with the emitted fluorescence. Standard filters can also be used for calibrating purposes. | |
| Flavin adenine dinucleotide | FAD, FADH2 | Flavin adenine dinucleotide, FAD and FADH2, is an oxidation-reduction prosthetic group (redox cofactor; compare NADH). FMN and FAD are the prosthetic groups of flavoproteins (flavin dehydrogenases). Type F substrates (fatty acids) generate FADH2, the substrate of electron transferring flavoprotein (CETF). Thus FADH2 forms a junction or funnel of electron transfer to CETF, the F-junction (compare N-junction, Q-junction), in the F-pathway control state. In contrast, FADH2 is not the substrate but the internal product of succinate dehydrogenase (CII). FAD is the oxidized (quinone) form, which is reduced to FADH2 (hydroquinone form) by accepting two electrons and two protons. |
| Flavonoids | Flavonoids are a group of bioactive polyphenols with potential antioxidant and anti-inflammatory effects, abundant in fruits and vegetables, and in some medicinal herbs. Flavonoids are synthesized in plants from phenylalanine. Dietary intake of flavonoids as nutraceuticals is discussed for targeting T2D and other degenerative diseases. | |
| Flow | I [MU∙s-1] | In an isomorphic analysis, any form of flow, I is the advancement of a process per unit of time, expressed in a specific motive unit [MU∙s-1], e.g., ampere for electric flow or current [A≡C∙s-1], watt for heat flow [W≡J∙s-1], and for chemical flow the unit is [mol∙s-1]. Flow is an extensive quantity. The corresponding isomorphic forces are the partial exergy (Gibbs energy) changes per advancement [J∙MU-1], expressed in volt for electric force [V≡J∙C-1], dimensionless for thermal force, and for chemical force the unit is [J∙mol-1], which deserves a specific acronym ([Jol]) comparable to volt. |
| Fluo calibration - DatLab | ||
| Fluorescence | Fluorescence is the name given to light emitted by a substance when it is illuminated (excited) by light at a shorter wavelength. The incident light causes an electron transition to a higher energy band in the molecules. The electron then spontaneously returns to its original energy state emitting a photon. The intensity of the emitted light is proportional to the concentration of the substance. Fluorescence is one form of Luminescence, especially Photoluminescence. | |
| Fluorescence-Control Unit | Fluorescence-Control Unit with O2k-Front Fixation, Current-Control (O2k-Chamber A and B) for regulation of light intensity of the LED in the fluorescence sensors. This item is a standard component of the O2k-Fluorescence LED2-Module. | |
| Fluorescence-Sensor Blue | Fluorescence-Sensor Blue: excitation LED 465 nm (dominant wavelength), photodiode, Filter-Cap equipped with Filter Set Saf for measurement of mitochondrial membrane potential with Safranin when delivered. The filter set Filter Set MgG / CaG for Magnesium green® / Calcium green® measurements is included. | |
| Fluorescence-Sensor Green | Fluorescence-Sensor Green: excitation LED 525 nm (dominant wavelength), photodiode, Filter-Cap equipped with Filter Set AmR for Amplex UltraRed measurements when delivered. | |
| Fluorescent marker | See Extrinsic fluorophores | |
| Fluorometric dyes | Extrinsic fluorophores; fluorescent markers. | |
| Fluorometry | Fluorometry (or fluorimetry) is the general term given to the method of measuring the fluorescent emission of a substance following excitation by light at a shorter wavelength. | |
| Fluorophore | A fluorophore is a fluorescent substance that may occur naturally (intrinsic fluorophores) or that may be added to a sample or preparation whereby the fluorescence intensity is proportional to the concentration of a specific species or parameter within the sample. These are extrinsic fluorophores, also referred to as fluorescent markers. | |
| Flux | J | Flux, J, is a specific quantity. Flux is flow, I [MU·s-1 per system] (an extensive quantity), divided by system size. Flux (e.g., oxygen flux) may be volume-specific (flow per volume [MU·s-1·L-1]), mass-specific (flow per mass [MU·s-1·kg-1]), or marker-specific (e.g. flow per mtEU). The motive unit [MU] of chemical flow or flux is the advancement of reaction [mol] in the chemical format. |
| Flux / Slope | J | Flux / Slope is the time derivative of the signal. In DatLab, Flux / Slope is the name of the pull-down menu for (1) normalization of flux (chamber volume-specific flux, sample-specific flux or flow, or flux control ratios), (2) flux baseline correction, (3) Instrumental background oxygen flux, and (4) flux smoothing, selection of the scaling factor, and stoichiometric normalization using a stoichiometric coefficient. Before changing the normalization of flux from volume-specific flux to sample-specific flux or flow, or flux control ratios, please be sure to use the standard Layout 04a (Flux per volume) or 04b (Flux per volume overlay). When starting with the instrumental standard Layouts 1-3, which display the O2 slope negative, the sample-specific flux or flow, or flux control ratios will not be automatically background corrected. To obtain the background corrected specific flux or flux control ratios, it is needed to tick the background correction in the lower part of the slope configuration window. Background correction is especially critical when performing measurements in a high oxygen regime or using samples with a low respiratory flux or flow. |
| Flux analysis - DatLab | The strategy of Flux analysis using DatLab depends on the research question and the corresponding settings applied in DatLab when recording the data with the O2k. Usng SUIT protocols, a sequence of respiratory steady-states is measured, marks are set, and numerical data are summarized in Mark statistics (F2). An AI approach is kept in mind when describing guidelines for evaluation of steady-states during data recording and analysis. | |
| Flux baseline correction | bc | Flux baseline correction provides the option to display the plot and all values of the flux (or flow, or flux control ratio) as the total flux, J, minus a baseline flux, J0.
JV(bc) = JV - JV0 JV = (dc/dt) · ν-1 · SF - J°VFor the oxygen channel, JV is O2 flux per volume [pmol/(s·ml)] (or volume-specific O2 flux), c is the oxygen concentration [nmol/ml = µmol/l = µM], dc/dt is the (positive) slope of oxygen concentration over time [nmol/(s · ml)], ν-1 = -1 is the stoichiometric coefficient for the reaction of oxygen consumption (oxygen is removed in the chemical reaction, thus the stoichiometric coefficient is negative, expressing oxygen flux as the negative slope), SF=1,000 is the scaling factor (converting units for the amount of oxygen from nmol to pmol), and J°V is the volume-specific background oxygen flux (Instrumental background oxygen flux). Further details: Flux / Slope. |
| Flux control efficiency | jZ-Y | Flux control efficiencies express the control of respiration by a metabolic control variable, X, as a fractional change of flux from YX to ZX, normalized for ZX. ZX is the reference state with high (stimulated or un-inhibited) flux; YX is the background state at low flux, upon which X acts.
Complementary to the concept of flux control ratios and analogous to elasticities of metabolic control analysis, the flux control efficiency of X upon background YX is expressed as the change of flux from YX to ZX normalized for the reference state ZX. » MiPNet article |
| Flux control ratio | FCR | Flux control ratios FCRs are ratios of oxygen flux in different respiratory control states, normalized for maximum flux in a common reference state, to obtain theoretical lower and upper limits of 0.0 and 1.0 (0 % and 100 %). For a given protocol or set of respiratory protocols, flux control ratios provide a fingerprint of coupling and substrate control independent of (1) mt-content in cells or tissues, (2) purification in preparations of isolated mitochondria, and (3) assay conditions for determination of tissue mass or mt-markers external to a respiratory protocol (CS, protein, stereology, etc.). FCR obtained from a single respirometric incubation with sequential titrations (sequential protocol; SUIT protocol) provide an internal normalization, expressing respiratory control independent of mitochondrial content and thus independent of a marker for mitochondrial amount. FCR obtained from separate (parallel) protocols depend on equal distribution of subsamples obtained from a homogenous mt-preparation or determination of a common mitochondrial marker. |
| Force | F; dmFX; ΔtrFX [J·MU-1] | Force is an intensive quantity. The product of force times advancement is the work (exergy) expended in a process or transformation. Force times flow is power [W].
|
| Forceps for membrane application | Forceps for membrane application: for OroboPOS and ISE membrane application; do not use for tissue preparation. | |
| Forceps\stainless Steel\angular Tip\fine | Forceps\stainless Steel\angular Tip\fine: for tissue preparation, stainless steel. Two pairs are used particularly for muscle fiber separation. | |
| Forceps\stainless Steel\rounded Tip\sharp | Forceps\stainless Steel\rounded Tip\sharp: for tissue preparation, stainless steel, antimagnetic. One pair is recommended for placing the tissue sample onto the microbalance and for handling in combination with Forceps\stainless Steel\straight Tip\sharp. | |
| Forceps\stainless Steel\straight Tip\sharp | Forceps\stainless Steel\straight Tip\sharp: for tissue preparation, stainless steel, antimagnetic. One pair is recommended for insertion of the sample into the O2k-chamber and for handling in combination with Forceps\stainless Steel\rounded Tip\sharp. | |
| Format | 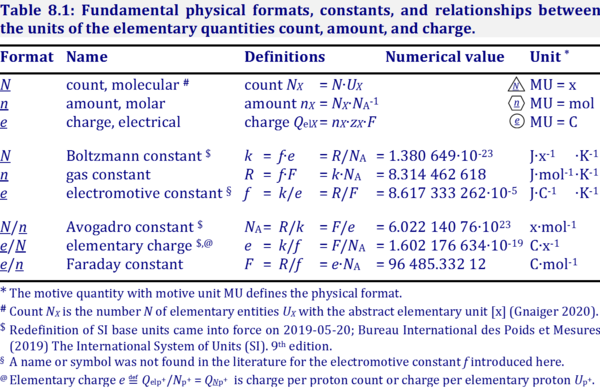 Converstion between different motive formats and corresponding motive units (Gnaiger 2020 BEC MitoPathways) Different formats can be chosen to express physicochemical quantities (motive entities or transformants) in corresponding motive units [MU]. Fundamental formats for electrochemical transformations are:
| |
| Free activity | αX [MU·m-3] | Free activity αX [MU·m-3] is pressure divided by isomorphic force. In the chemical amount format, αX is expressed in units of concentration of X [mol·L-1]. αX is the local concentration in a concentration gradient. If the concentration gradient is collapsed to a boundary of zero thickness in a compartmental system, αX reflects the singularity in the transition between the two phases or compartments. |
| Free radicals | A free radical is any atom or molecule that contains one or more unpaired electrons in an orbital. The degree of chemical reactivity depends on the localization of unpaired electrons. Free radicals are extremely reactive, and they can either donate or accept an electron from other molecules. Free radicals that include oxygen radicals and derivatives of oxygen are reactive oxygen species (ROS). Likewise, reactive nitrogen species (RNS) are nitric oxide-derived compounds. ROS/RNS include oxygen/nitrogen free radicals and non-radicals that are easily converted into radicals. Mitochondria are a main endogenous source of free radicals in cells and consequently are exposed to oxidative-nitrosative damage. Electron transfer in the electron transfer-pathway (ET-pathway) is not perfect, leading an electron leakage. This electron leakage permits the formation of ROS such as superoxide anion (O2•−), hydrogen peroxide (H2O2) and the hydroxyl radical (HO•). | |
| French Group of Bioenergetics | FGoB | The French Group of Bioenergetics... |
| Full screen | By clicking/enabling Full screen in the Graph-menu in DatLab the currently selected graph is shown alone on the full screen (On) or together with the other defined graphs (Off). Full screen is particularly useful for a single channel overview and for Copy to clipboard [ALT+G B]. | |
| Fumarase | FH | Fumarase or fumarate hydratase (FH) is an enzyme of the tricarboxylic acid cycle catalyzing the equilibrium reaction between fumarate and malate. Fumarase is found not only in mitochondria, but also in the cytoplasm of all eukaryotes. |
| Fura2 | Fura2 is a ratiometric fluorescence probe for the measurement of calcium. Its derivative Fura-2-acetoxymethyl ester (Fura2-AM) is membrane permable and can thus be used to measure intracellular free calcium concentration (Grynkiewicz et al., 1985). For this purpose, cells are incubated with Fura2-AM, which crosses the cell membrane by diffusion and is cleaved into free Fura2 and acetoxymethyl groups by cellular esterases. Intracellular free calcium is measured by exciting the dye at 340 nm and 380 nm, which are the excitation optima of calcium-bound and free Fura2, respectively, and emission detection above 500 nm. Through the ratiometric detection unequal distribution of the dye within the cell and other potential disturbances are largely cancelled out, making this a widely used and relatively reliable tool for calcium measurements. | |
| GM-pathway control state | GM | GM: Glutamate & Malate.
MitoPathway control state: NADH electron transfer-pathway state The GM-pathway control state (glutamate-malate pathway control state) is established when glutamate&malate are added to isolated mitochondria, permeabilized cells and other mitochondrial preparations. Glutamate and transaminase are responsible for the metabolism of oxaloacetate, comparable to the metabolism with acetyl-CoA and citrate synthase. |
| GMS-pathway control state | GMS | GMS: Glutamate & Malate & Succinate.
MitoPathway control: NS Transaminase catalyzes the reaction from oxaloacetate to 2-oxoglutarate, which then establishes a cycle without generation of citrate. OXPHOS is higher with GS (CI&II) compared to GM (CI) or SRot (CII). This documents an additive effect of convergent CI&II electron flow to the Q-junction, with consistent results obtained with permeabilized muscle fibres and isolated mitochondria (Gnaiger 2009). |
| Gain | The gain is an amplification factor applied to an input signal to increase the output signal. | |
| Gas constant | R [J·mol-1·K-1] | The gas constant, R = 8.314462618 J·mol-1·K-1, has the SI unit for energy per amount per temperature. R is primarily known from the ideal gas equation, pV = nRT or p = cRT. Therefore, RT is the ratio of pressure p and concentration c.
R = f·F, the electrochemical constant f times the Faraday constant F. R = k·NA, the Boltzmann constant k times the Avogadro constant NA. |
| Getting started - DatLab | Users have to enter their user details the first time they use DatLab 8 on a specific computer. As well, entering some basic settings is required when connecting DatLab 8 with an O2k for the first time. | |
| Gibbs energy | G [J] | Gibbs energy G [J] is exergy which cannot be created internally (subscript i), but in contrast to internal-energy (diU/dt = 0) is not conserved but is dissipated (diG/dt < 0) in irreversible energy transformations at constant temperature and (barometric) pressure, T,p. Exergy is available as work in reversible energy transformations (100 % efficiency), and can be partially conserved when the exergonic transformation is coupled to an endergonic transformation. |
| Glucose | Glc | Glucose, also known as D-glucose or dextrose, is a monosaccharide and an important carbohydrate in biology. Cells use it as the primary source of energy and a metabolic intermediate. |
| Glutamate | G | Glutamic acid, C5H9NO4, is an amino acid which occurs under physiological conditions mainly as the anion glutamate-, G, with pKa1 = 2.1, pKa2 = 4.07 and pKa3 = 9.47. Glutamate&malate is a substrate combination supporting an N-linked pathway control state, when glutamate is transported into the mt-matrix via the glutamate-aspartate carrier and reacts with oxaloacetate in the transaminase reaction to form aspartate and oxoglutarate. Glutamate as the sole substrate is transported by the electroneutral glutamate-/OH- exchanger, and is oxidized in the mitochondrial matrix by glutamate dehydrogenase to α-ketoglutarate (2-oxoglutarate), representing the glutamate-anaplerotic pathway control state. Ammonia (the byproduct of the reaction) passes freely through the mitochondrial membrane. |
| Glutamate dehydrogenase | mtGDH | Glutamate dehydrogenase, located in the mitochondrial matrix (mtGDH), is an enzyme that converts glutamate to α-ketoglutarate [1]. mtGDH is not part of the TCA cycle, but is involved in glutaminolysis as an anaplerotic reaction. |
| Glutamate-anaplerotic pathway control state | G | G: Glutamate is an anaplerotic NADH-linked type 4 substrate (N). When supplied as the sole fuel substrate in the glutamate-anaplerotic pathway control state, G is transported by the electroneutral glutamate-/OH- exchanger, and is oxidised via mt-glutamate dehydrogenase in the mitochondrial matrix. The G-pathway plays an important role in glutaminolysis. |
| Glutamate-aspartate carrier | The glutamate-aspartate carrier catalyzes the electrogenic antiport of glutamate- +H+ for aspartate-. It is an important component of the malate-aspartate shuttle in many mitochondria. Due to the symport of glutamate- + +H+, the glutamate-aspartate antiport is not electroneutal and may be impaired by uncoupling. Aminooxyacetate is an inhibitor of the glutamate-aspartate carrier. | |
| Glycerophosphate | Gp | Glycerophosphate (synonym: α-glycerophosphate; glycerol-3-phosphate; C3H9O6P) is an organophosphate and it is a component of glycerophospholipids. The mitochondrial Glycerophosphate dehydrogenase Complex oxidizes glycerophosphate to dihydroxyacetone phosphate and feeds electrons directly to ubiquinone. |
| Glycerophosphate dehydrogenase Complex | CGpDH | Glycerophosphate dehydrogenase complex (CGpDH) is a Complex of the electron transfer-pathway localized at the outer face of the mt-inner membrane. CGpDH is thus distinguished from cytosolic GpDH. CGpDH oxidizes glycerophosphate to dihydroxyacetone phosphate and feeds two electrons into the Q-junction, thus linked to an ET pathway level 3 control state. |
| Glycerophosphate pathway control state | Gp | The glycerophosphate pathway control state (Gp) is an ET-pathway level 3 control state, supported by the fuel substrate glycerophosphate and electron transfer through glycerophosphate dehydrogenase Complex into the Q-junction. The glycerolphosphate shuttle represents an important pathway, particularly in liver and blood cells, of making cytoplasmic NADH available for mitochondrial oxidative phosphorylation. Cytoplasmic NADH reacts with dihydroxyacetone phosphate catalyzed by cytoplasmic glycerophos-phate dehydrogenase. On the outer face of the inner mitochondrial membrane, mitochondrial glycerophosphate dehydrogenase oxidises glycerophosphate back to dihydroxyacetone phosphate, a reaction not generating NADH but reducing a flavin prosthesic group. The reduced flavoprotein donates its reducing equivalents to the electron transfer-pathway at the level of CoQ. |
| Glycerophosphate shuttle | Gp shuttle | The glycerophosphate shuttle makes cytoplasmic NADH available for mitochondrial oxidative phosphorylation. Cytoplasmic NADH reacts with dihydroxyacetone phosphate catalyzed by cytoplasmic glycerophosphate dehydrogenase. On the outer face of the inner mitochondrial membrane, glycerophosphate dehydrogenase complex (mitochondrial glycerophosphate dehydrogenase) oxidizes glycerophosphate back to dihydroxyacetone phosphate, a reaction not generating NADH but reducing a flavin prosthesic group. The reduced flavoprotein transfers its reducing equivalents into the Q-junction, thus representing a ET pathway level 3 control state. |
| Gnaiger 2019 MiP2019 | ||
| Graph control - DatLab | A combination of mouse and keyboard commands provides convenient control of graphs in DatLab 8. | |
| Graph layout - DatLab | » See Layout for DatLab graphs. | |
| Graph options - DatLab | Several display options can be applied to a DatLab graph under Graph options. | |
| Group | See population. | |
| H2DCFDA | DCF, H2-DCF | H2DCFDA (dichlorodihydrofluorescein diacetate) is a cell permeant fluorescent probe that has been used as an indicator of ROS presence. It is a reduced form of fluorescein that does not present fluorescence. After entry in the cell, it suffers deacetylation by intracellular esterases, and upon oxidation it is converted to dichlorofluorescein (excitation wavelength ~492–495 nm, emission ~517–527 nm). It may be oxidised by hydrogen peroxide, hydroxyl radical, hypochlorite anion, nitric oxide, peroxyl radical, peroxynitrite, singlet oxygen and superoxide. Has been used as a general indicator of ROS by fluorescence microscopy. |
| HPTS | HPTS | 8-Hydroxypyrene-1,3,6-trisulfonic acid trisodium salt (HPTS) is a ratiometric pH fluorophore; pKa = 7.3. Relative molecular mass: Mr = 524.39 |
| Harmonization | Harmonization is the process of minimizing redundant or conflicting standards which may have evolved independently. To obtain a common basis in reaching a defined objective, critical requirements are identified that need to be retained. | |
| Harmonized European norm | EN-norm | Harmonized European norms are norms valid for all members of the European Union. They are mandatory parts of the individual national collections of norms. |
| Harmonized SUIT protocols | H-SUIT | Harmonized SUIT protocols (H-SUIT) are designed to include cross-linked respiratory states. When performing harmonized SUIT protocols in parallel, measurements of cross-linked respiratory states can be statistically evaluated as replicates across protocols. Additional information is obtained on respiratory coupling and substrate control by including respiratory states that are not common (not cross-linked) across the harmonized protocols. |
| Harmonized standard | A harmonized standard is a European standard developed by a recognized European Standards Organisation: CEN, CENELEC, or ETSI. | |
| Healthy ageing | Healthy ageing: 'WHO has released the first World report on ageing and health, reviewing current knowledge and gaps and providing a public health framework for action. The report is built around a redefinition of healthy ageing that centres on the notion of functional ability: the combination of the intrinsic capacity of the individual, relevant environmental characteristics, and the interactions between the individual and these characteristics' (Beard 2016 The Lancet). | |
| Healthy reference population | HRP | A healthy reference population, HRP, establishes the baseline for the relation between body mass and height in healthy people of zero underweight or overweight, providing a reference for evaluation of deviations towards underweight or overweight and obesity. The WHO Child Growth Standards (WHO-CGS) on height and body mass refer to healthy girls and boys from Brazil, Ghana, India, Norway, Oman and the USA. The Committee on Biological Handbooks compiled data on height and body mass of healthy males from infancy to old age (USA), published before emergence of the fast-food and soft-drink epidemic. Four allometric phases are distinguished with distinct allometric exponents. At heights above 1.26 m/x the allometric exponent is 2.9, equal in women and men, and significantly different from the exponent of 2.0 implicated in the body mass index, BMI [kg/m2]. |
| Heat | Q, Qth [J] | Heat is a form of energy [J]. The relationship between heat and work provides the foundation of thermodynamics, which describes transformations from an initial to a final state of a system. In energy transformations heat may pass through the boundary of the system, at an external heat flow of deQ/dt. |
| Height of humans | h [m]; H [m·x-1] | The height of humans, h, is given in SI units in meters [m]. Humans are countable objects, and the symbol and unit of the number of objects is N [x]. The average height of N objects is, H = h/N [m/x], where h is the heights of all N objects measured on top of each other. Therefore, the height per human has the unit [m·x-1] (compare body mass [kg·x-1]). Without further identifyer, H is considered as the standing height of a human, measured without shoes, hair ornaments and heavy outer garments. |
| Heterothermy | Heterothermy is the variable regulation of body temperature in endotherms which can change their body temperatures as levels of activity and environmental conditions dictate (e.g. hibernators). In regional heterothermy, temperature gradients are present, e.g. between body core and extremeties. | |
| Hexokinase | HK | The hexokinase catalyzes the phosphorylation of D-glucose at position 6 by ATP to yield D-glucose 6-phosphate as well as the phosphorylation of many other hexoses like D-fructose, D-mannose, D-glucosamine. |
| Heymsfield 2014 Am J Clin Nutr | ||
| High signal at zero oxygen | A high signal at zero oxygen may be observed during zero calibration (R0). First, check the quality of the dithionite solution. The following instructions show how to distinguish between a defective sensor head and an electrical leak current. | |
| High-resolution respirometry | HRR | High-resolution respirometry, HRR, is the state-of-the-art approach in mitochondria and cell research to measure respiration in various types of mitochondrial preparations and living cells combined with MultiSensor modules.
Mitochondrial function and dysfunction have gained increasing interest, reflecting growing awareness of the fact that mitochondria play a pivotal role in human health and disease. HRR combines instrumental accuracy and reliability with the versatility of applicable protocols, allowing practically unlimited addition and combination of substrates, inhibitors, and uncouplers using the Oroboros O2k-technology. Substrate-uncoupler-inhibitor titration (SUIT) protocols allow the interrogation of numerous mitochondrial pathway and coupling states in a single respirometric assay. Mitochondrial respiratory pathways may be analyzed in detail to evaluate even minor alterations in respiratory coupling and pathway control patterns. The O2k-technology provides sole source instruments, with no other available instrument meeting its specifications for high-resolution respirometry. Technologically, HRR is based on the Oroboros O2k-technology, combining optimized chamber design, application of oxygen-tight materials, electrochemical sensors, Peltier-temperature control, and specially developed software features (DatLab) to obtain the unique sensitive and quantitative resolution of oxygen concentration and oxygen flux, with both, a closed-chamber or open-chamber mode of operation (TIP2k). Standardized calibration of the polarographic oxygen sensor (static sensor calibration), calibration of the sensor response time (dynamic sensor calibration), and evaluation of instrumental background oxygen flux (systemic flux compensation) provide the experimental basis for high accuracy of quantitative results and quality control in HRR. HRR can be extended for MultiSensor analysis by using the O2k-Fluo Smart-Module. Smart Fluo-Sensors are integrated into the O2k to measure simultaneously fluorometric signals using specific fluorophores. Potentiometric modules are available with ion-selective electrodes (pH, TPP+). The PB-Module extends HRR to PhotoBiology with accurate control of the light intensity and measurement of photosynthesis. The O2k and the NextGen-O2k support all these O2k-Modules. The NextGen-O2k all-in-one, however, is unique in supporting Q-Redox and NADH-Redox Modules. |
| Holode | Small entetic units are counted into the reference system on a balance opposite to the experimental system with the large sample, which in balance contains as many abstract units as the count of entetic units in the reference system. | |
| Homeothermy | Homeothermy is the stable regulation of body temperature in endotherms by metabolic heat production and control of heat exchange with the environment, or in ectotherms by behavioural means to select a stable thermal environment. | |
| Hood 2019 Nutr Diabetes | ||
| Horseradish peroxidase | HRP | Horseradish peroxidase readily combines with hydrogen peroxide (H2O2) and the resultant [HRP-H2O2] complex can oxidize a wide variety of hydrogen donors. |
| Hydride | H- | The hydride anion is the species H−. |
| Hydrogen | H2 | Molecular hydrogen H2 is a constituent of the air with a volume fraction of 0.00005. It is a colorless and odorless gas with a molecular mass of 2.016. Its pharmacological potential and effects on mitochondrial metabolism are discussed in various publications without complete evidence on the underlying mechanisms. |
| Hydrogen ion | H+ | The terms hydrogen ion H+ and proton, p or p+, are used synonymously in chemistry. A hydrogen ion is a positively charged molecule. In particle physics, however, a proton is a submolecular and subatomic particle with a positive electric charge. The H+ ion has no electrons and is a bare charge with only about 1/64 000 of the radius of a hydrogen atom. Free H+ is extremely reactive, with an extremely short lifetime in aqueous solutions. There H+ forms the hydronium ion H3O+, which in turn is further solvated by water molecules in clusters such as H5O2+ and H9O4+. The transfer of H+ in an acid–base reaction is referred to as proton transfer. The acid is the H+ donor and the base is the H+ acceptor. |
| Hydrogen ion pump | Mitochondrial hydrogen ion pumps — frequently referred to as "proton pumps" — are large enzyme complexes (CI, CIII, CIV, ATP synthase) spanning the mt-inner membrane mtIM, partially encoded by mtDNA. CI, CIII and CIV are H+ pumps that drive hydrogen ions against the electrochemical protonmotive force pmF and thus generating the pmF, driven by electron transfer from reduced substrates to oxygen. In contrast, ATP synthase (also known as CV) is a H+ pump that utilizes the exergy of proton flow along the protonmotive force to drive phosphorylation of ADP to ATP. | |
| Hydrogen peroxide | H2O2 | Hydrogen peroxide, H2O2 or dihydrogen dioxide, is one of several reactive oxygen intermediates generally referred to as reactive oxygen species (ROS). It is formed in various enzyme-catalyzed reactions (e.g., superoxide dismutase) with the potential to damage cellular molecules and structures. H2O2 is dismutated by catalase to water and oxygen. H2O2 is produced as a signaling molecule in aerobic metabolism and passes membranes more easily compared to other ROS. |
| Hydrogen sulfide | H2S | Hydrogen sulfide (H2S) is involved in signaling and may have have further biological importance. |
| Hydrogenion flux | JH+ | Volume-specific hydrogenion flux or H+ flux is measured in a closed system as the time derivative of H+ concentration, expressed in units [pmol·s-1·mL-1]. H+ flux can be measured in an open system at steady state, when any acidification of the medium is compensated by external supply of an equivalent amount of base. The extracellular acidification rate (ECAR) is the change of pH in the incubation medium over time, which is zero at steady state. Volume-specific H+ flux is comparable to volume-specific oxygen flux [pmol·s-1·mL-1], which is the (negative) time derivative of oxygen concentration measured in a closed system, corrected for instrumental and chemical background. pH is the negative logarithm of hydrogen ion activity. Therefore, ECAR is of interest in relation to acidification issues in the incubation buffer or culture medium. The physiologically relevant metabolic H+ flux, however, must not be confused with ECAR. |
| Hydron | H+ | Hydron is the general name for the cation H+ used without regard to the nuclear mass of the hydrogen entity (H is the hydro group), either for H in its natural abundance or without distinction between the isotopes. |
| Hydronium ion | H3O+ | H+ forms the hydronium ion H3O+, which in turn is further solvated by water molecules in clusters such as H5O2+ and H9O4+. |
| Hydroxybutyrate | β-hydroxybutyrate or 3-hydroxybutyrate is a ketone body that can be used as a NADH-linked substrate. The β-hydroxybutyrate dehydrogenase produces acetoacetate while reducing NAD+ to NADH.
| |
| Hydroxycinnamate | Hci | Hydroxycinnamate (alpha-cyano-4-hydroxycinnamic acid) is an inhibitor of the pyruvate carrier (0.65 mM). Above 10 mM pyruvate, hydroxycinnamate cannot inhibit respiration from pyruvate, since the weak pyruvic acid can pass the inner mt-membrane in non-dissociated form. |
| Hydroxylamine | Hydroxylamine is an inhibitor of catalase. | |
| Hyperoxia | hyperox | Hyperoxia is defined as environmental oxygen pressure above the normoxic reference level. Cellular and intracellular hyperoxia is imposed on isolated cells and isolated mitochondria at air-level oxygen pressures which are higher compared to cellular and intracellular oxygen pressures under tissue conditions in vivo. Hyperoxic conditions may impose oxidative stress and may increase maximum aerobic performance. |
| Hyperthermia | Hyperthermia in endotherms is a state of stressful up to lethal elevated body core temperature. In humans, the limit of hyperthermia (fever) is considered as >38.3 °C, compared to normothermia at a body temperature of 36.5 to 37.5 °C. | |
| Hyphenation | Hyphenation is used to connect two words (compound words) or two parts of a word to clarify the meaning of a sentence. The same two words may be hyphenated or not depending on context. Hyphenation may present a problem when searching for a term such as 'Steady state'. It is helpful to write 'steady-state measurement', to clarify that the measurement is performed at steady state, rather than implying that a state measurement is steady. But this does not imply that hyphenation is applied to the 'measurement performed at steady state'. Thus, the key word is 'steady state'. Compound adjectives should be hyphenated (steady-state measurement), but if the compound adjective follows the term (measurement at steady state), hyphenation does not add any information and should be avoided. Find more examples and guidelines in the grammarly blog on Hyphen and in apastyle.apa.org. | |
| Hypothermia | Hypothermia in endotherms is a state of stressful up to lethal low body core temperature. In humans, the limit of hypothermia is considered as 35 °C, compared to normothermia at a body temperature of 36.5 to 37.5 °C. Hypothermia is classified as mild (32–35 °C), moderate (28–32 °C), severe (20–28 °C), and profound (<20 °C). | |
| Hypoxia | hypox | Hypoxia (hypox) is defined in respiratory physiology as the state when insufficient O2 is available for respiration, compared to environmental hypoxia defined as environmental oxygen pressures below the normoxic reference level. Three major categories of hypoxia are (1) environmental hypoxia, (2) physiological tissue hypoxia in hyperactivated states (e.g. at VO2max) with intracellular oxygen demand/supply balance at steady state in tissues at environmental normoxia, compared to tissue normoxia in physiologically balanced states, and (3) pathological tissue hypoxia including ischemia and stroke, anaemia, chronic heart disease, chronic obstructive pulmonary disease, severe COVID-19, and obstructive sleep apnea. Pathological hypoxia leads to tissue hypoxia and heterogenous intracellular anoxia. Clinical oxygen treatment ('environmental hyperoxia') may not or only partially overcome pathological tissue hypoxia. |
| IRDiRC | IRDiRC |  The International Rare Diseases Research Consortium (IRDiRC) teams up researchers and organizations investing in rare diseases research in order to achieve two main objectives by the year 2020, namely to deliver 200 new therapies for rare diseases and means to diagnose most rare diseases. The International Rare Diseases Research Consortium (IRDiRC) teams up researchers and organizations investing in rare diseases research in order to achieve two main objectives by the year 2020, namely to deliver 200 new therapies for rare diseases and means to diagnose most rare diseases. |
| ISE Package 1 TPP or Ca | O2k-TPP+ and Ca2+ ISE\1 Chamber: ISE-Package for 1 TPP+ and Ca2+ electrode. | |
| ISE-Ca2+ Membranes | ISE-Ca2+ Membranes: PVC, 4 mm diameter, box of 5 membranes. To be used with the O2k-TPP+ ISE-Module. | |
| ISE-Compressible Tube | ISE-Compressible Tube for Ion-Selective Electrode TPP+ and Ca2+. | |
| ISE-Filling Syringe | ISE-Filling Syringe with needle for Ion-Selective Electrode TPP+ and Ca2+. | |
| ISE-Inner Glass Electrode | ISE-Inner Glass Electrode of ISE, with Ag/AgCl- and Pt-wire | |
| ISE-Membrane Mounting Tool | ISE-Membrane Mounting Tool for Ion-Selective Electrode TPP+ and Ca2+. O2k-TPP+ ISE-Module: mounting tool included. | |
| ISE-Membrane Seal | 'ISE-Membrane Seal for Ion-Selective Electrode TPP+ and Ca2+. | |
| ISE-TPP+ Membranes | ISE-TPP+ Membranes, PVC, 4 mm diameter, box of 5 membranes. | |
| ISO 10012:2003 Measurement management systems | ISO 10012:2003 | ISO 10012:2003 Measurement management systems — Requirements for measurement processes and measuring equipment: An effective measurement management system ensures that measuring equipment and measurement processes are fit for their intended use and is important in achieving product quality objectives and managing the risk of incorrect measurement results. The objective of a measurement management system is to manage the risk that measuring equipment and measurement processes could produce incorrect results affecting the quality of an organization’s product. The methods used for the measurement management system range from basic equipment verification to the application of statistical techniques in the measurement process control. |
| ISO 13528:2015 Statistical methods for use in proficiency testing by interlaboratory comparison | ISO 13528:2015 | ISO 13528:2015 Statistical methods for use in proficiency testing by interlaboratory comparison: Proficiency testing involves the use of interlaboratory comparisons to determine the performance of participants (which may be laboratories, inspection bodies, or individuals) for specific tests or measurements, and to monitor their continuing performance. There are a number of typical purposes of proficiency testing ISO/IEC 17043:2010. These include the evaluation of laboratory performance, the identification of problems in laboratories, establishing effectiveness and comparability of test or measurement methods, the provision of additional confidence to laboratory customers, validation of uncertainty claims, and the education of participating laboratories. The statistical design and analytical techniques applied must be appropriate for the stated purpose(s). |
| ISO 15189:2012 Medical laboratories — Particular requirements for quality and competence | ISO 15189:2012 | ISO 15189:2012 Medical laboratories — Particular requirements for quality and competence: This International Standard is for use by medical laboratories in developing their quality management systems and assessing their own competence, and for use by accreditation bodies in confirming or recognising the competence of medical laboratories. While this International Standard is intended for use throughout the currently recognised disciplines of medical laboratory services, those working in other services and disciplines could also find it useful and appropriate. |
| ISO 17511:2003 In vitro diagnostic medical devices | ISO 17511:2003 | ISO 17511:2003 In vitro diagnostic medical devices -- Measurement of quantities in biological samples -- Metrological traceability of values assigned to calibrators and control materials: For measurements of quantities in laboratory medicine, it is essential that the quantity is adequately defined and that the results reported to the physicians or other health care personel and patients are adequately accurate (true and precise) to allow correct medical interpretation and comparability over time and space. |
| ISO 9001:2015 Quality management systems - requirements | ISO 9001:2015 | ISO 9001:2015 Quality management systems - requirements: The adoption of a quality management system is a strategic decision for an organization that can help to improve its overall performance and provide a sound basis for sustainable development initiatives. Consistently meeting requirements and addressing future needs and expectations poses a challenge for organizations in an increasingly dynamic and complex environment. To achieve this objective, the organization might find it necessary to adopt various forms of improvement in addition to correction and continual improvement, such as breakthrough change, innovation and re-organization. |
| ISO/IEC 17025:2005 Competence of testing and calibration laboratories | ISO/IEC 17025:2005 | ISO/IEC 17025:2005 General requirements for the competence of testing and calibration laboratories: The use of this International Standard will facilitate cooperation between laboratories and other bodies, and assist in the exchange of information and experience, and in the harmonization of standards and procedures. This International Standard specifies the general requirements for the competence to carry out tests and/or calibrations, including sampling. It covers testing and calibration performed using standard methods, non-standard methods, and laboratory-developed methods. |
| ISO/IEC 17043:2010 General requirements for proficiency testing | ISO/IEC 17043:2010 | ISO/IEC 17043:2010 Conformity assessment — General requirements for proficiency testing: The use of interlaboratory comparisons is increasing internationally. This International Standard provides a consistent basis to determine the competence of organizations that provide proficiency testing. |
| ISS-Filter and Tubing | ISS-Filter and Tubing, ISS-Integrated Suction System. | |
| ISS-Integrated Suction System | ISS-Integrated Suction System: Suction pump with stainless steel housing, 2 liter waste bottle, filter and tubing; for siphoning off excess medium from the O2k-Stopper and for emptying the O2k-chambers. The ISS is included as a standard component of the O2k-FluoRespirometer. Media containing living cells or microorganisms, various poisons (inhibitors, uncouplers) and mixtures of proteins and substrates are safely disposed off in the 2-litre waste bottle. | |
| ISS-Lid | ISS-Lid, for ISS-Waste Bottle, component of the ISS-Integrated Suction System. | |
| ISS-Steel Housing | ISS-Steel Housing, a component of the ISS-Integrated Suction System. | |
| ISS-Waste Bottle | ISS-Waste Bottle, 2-liter, component of the ISS-Integrated Suction System. | |
| Iconic symbols | Iconic symbols are used in ergodynamics to indicate more explicitely — compared to standard SI or IUPAC symbols — the quantity represented and some boundary conditions. This is particularly the case in normalized quantities (ratios of quantities). Iconic (or canonical) symbols help to clarify the meaning, are based on SI and IUPAC symbols as far as possible, and may be translated into more commonly used, practical symbols. Several ambiguities in SI and IUPAC symbols are eliminated by the systematic structure of iconic symbols, but it may be impossible to avoid all ambiguities, particulary when long (canonical) symbols are abbreviated in a particular context. Clarity is improved always by showing the unit of a quantity together with the symbol of the quantity. Iconic symbols cannot be identical with IUPAC symbols when a different definition is used — this would add to the confusion. For example, the IUPAC symbols nB [mol] and VB [m3] denote amount and volume of B. Consequently, it should be expected, that the symbol QB indicates charge of B [C]. However, the IUPAC symbol QB is used for particle charge per ion B [C·x-1]. This prohibits a consistent definition of QB as a potential iconic symbol for charge carried by a given quantity of ions B with unit [C], instead of particle charge per ion B with unit [C·x-1]. Hence, the conventional ambigous system forces compatible iconic symbols to be more complicated, using QelB [C] and QNB [C·x-1] to distinguish charge of B from charge per elementary B. QnB [C·mol-1] is charge per molar amount of B. | |
| Illumination | F10 | The chambers of the Oroboros O2k are illuminated by an internal LED. The illumination is switched on and off in DatLab during the experiment by pressing [F10]. This illumination must be distinguished from light introduced into the chambers by LEDs for the purpose of spectrophotometric and fluorometric measurements. For these, the internal illumination must be switched off. |
| Illumination on/off | F10 | The illumination in both chambers is switched on/off. |
| Impact factor | IF | Impact factor is a measure of a scientific journal's citations per publication. The Journal Citation Reports, maintained by Clarivate Analytics, provides the calculated impact factors. The IF is frequently used as an indicator of a journal's importance or prestige, which is nowadays increasingly contested. |
| Improvement score | RIS | The relative improvement score, RIS, provides a measure of improvement of a trait from a value measured at baseline, B, to a value measured after treatment, T, expressing the total improvement, T-B, in relation to the theoretical scope of improvement and the level of the trait observed at baseline. RIS incorporates the concept of diminishing returns and consideres maintaining a high value of a trait as an improvement relative to the potential loss. |
| In vitro diagnostic medical device | IVD | A medical device is an in vitro diagnostic medical device (IVD) if it is a reagent, calibrator, control material, kit, specimen receptacle, software, instrument, apparatus, equipment or system, whether used alone or in combination with other diagnostic goods for in vitro use. |
| Incident light | The term incident light is used for a beam of light falling upon a surface. | |
| Inclusion criteria | The Inclusion criteria are based on key features of the target population that the researchers will use to answer their question. These criteria should identify the study population in a consistent, reliable, uniform, and objective manner. With the Exclusion criteria, this factor must be a cofounder for the outcome parameter | |
| Indian Academy of Pediatrics Growth Charts Committee 2015 Indian Pediatr | ||
| Inorganic phosphate | Pi | Inorgnic phosphate (Pi) is a salt of phosphoric acid. In solution near physiological pH, the species HPO42- and H2PO4- dominate. See also: Phosphate carrier (Pic). |
| Inside the O2k | A glance inside the Oroboros O2k | |
| Install Oroboros protocol package | The standard Instrumental and SUIT DL-Protocols package is automatically implemented with the simple DatLab programme installation. We recommend a 'clean install': rename your previous DatLab programme subdirectory (e.g. C:\DatLab_OLD). Updates and newly developed DL protocols can be simply downloaded by clicking on [Protocols]\Install Oroboros protocol package. | |
| Instrumental: Browse DL-Protocols and templates | Instrumental DL-Protocols (DLP) including DatLab example traces, instructions, brief explanatory texts, links to relevant pages and templates for data evaluation can be browsed from inside DatLab 7.4. Click on menu [Protocols]\Instrumental: Browse DL-Protocols and templates to open a folder with all the DL-Protocols and templates for cleaning, calibration, and background determination provided with the DatLab 7.4. Select a sub-directory and open an DL-Protocol and/or template as desired. | |
| Integration time | Integration time is the time taken to scan a single full range spectrum using photodiode arrays. It is equivalent to the exposure time for a camera. The shortest integration time defines the fastest response time of a spectrophotometer. Increasing the integration time increases the sensitivity of the device. The white balance or balance and subsequent measurements must always be carried out at the same integration time. | |
| Intensive quantity | Intensive quantities are partial derivatives of an extensive quantity by the advancement, dtrξX, of an energy transformation tr; example: Force. In contrast to extensive quantities which pertain to the entire system and are additive, extensive quantities 'take well defined values at each point of the system' (Prigogine 1967 Interscience) and are non-additive. Intensive and extensive quantities can be easily discriminated by the units, e.g. [J] for the extensive quantity, in contrast to [J·mol-1] for the corresponding intensive quantity. In the general definition of thermodynamics, intensive quantities are not distinguished from specific quantities (Cohen 2008 IUPAC Green Book). Ergodynamics emphasizes the contrast between specific quantities which are extensive quantities normalized for a variable expressing system size (mass, volume of the system, amount of substance in a system) and intensive quantities which are normalized for the motive unit of a transformation (mass exchanged, volume change of the system, amount of substance reacting in a system; Gnaiger 1993 Pure Appl Chem). Intensive and specific quantities are both non-additive, take well defined values at each point of the system, and both corresponding quantities are expressed in identical units, e.g. the intensive quantity Gibbs force of a catabolic reaction (such as oxidation; 0 = -1 Glc - 6 O2 + 6 CO2 + 6 H2O), ΔkGGlc [kJ·mol-1], and the specific quantity Gibbs energy per mole glucose contained in a system, GGlc [kJ·mol-1] (with respect to an arbitrarily defined reference state, such as the reference state of formation or combustion). | |
| Interlaboratory comparison | An interlaboratory comparison is the organization, performance and evaluation of measurements or tests on the same or similar items by two or more laboratories in accordance with predetermined conditions. | |
| Internal flow | Ii [MU·s-1] | Within the system boundaries, irreversible internal flows, Ii,—including chemical reactions and the dissipation of internal gradients of heat and matter—contribute to internal entropy production, diS/dt. In contrast, external flows, Ie, of heat, work, and matter proceed reversibly across the system boundaries (of zero thickness). Flows are expressed in various formats per unit of time, with corresponding motive units [MU], such as chemical [mol], electrical [C], mass [kg]. Flow is an extensive quantity, in contrast to flux as a specific quantity. |
| Internal-energy | U [J] | Internal-energy, U [J], can neither be destroyed nor created (first law of thermodynamics: diU/dt = 0). Note that internal (subscript i), as opposed to external (subscript e), must be distinguished from "internal-energy", U, which contrasts with "Helmholtz energy", A, as enthalpy, H, contrasts with Gibbs energy, G. |
| International Mito Patients (IMP) | IMP |  The International Mito Patients is a network of national patient organizations involved in mitochondrial disease. Mitochondrial disease is a rare disease with a limited number of patients per country. The national patient organizations which are a member of IMP each are active and powerful in their own countries. By joining forces IMP can represent a large group of patients and as such be their voice on an international level. The International Mito Patients is a network of national patient organizations involved in mitochondrial disease. Mitochondrial disease is a rare disease with a limited number of patients per country. The national patient organizations which are a member of IMP each are active and powerful in their own countries. By joining forces IMP can represent a large group of patients and as such be their voice on an international level. |
| International Society for Mountain Medicine | ISMM | The International Society for Mountain Medicine is an interdisciplinary society comprising about xx members worldwide. Its purpose is .. |
| International Society on Oxygen Transport to Tissue | ISOTT | The International Society on Oxygen Transport to Tissue is an interdisciplinary society comprising about 250 members worldwide. Its purpose is to further the understanding of all aspects of the processes involved in the transport of oxygen from the air to its ultimate consumption in the cells of the various organs of the body. Founded in 1973, the society has been the leading platform for the presentation of many of the technological and conceptual developments within the field both at the meetings themselves and in the proceedings of the society. |
| International Standard Serial Number | ISSN | The International Standard Serial Number, ISSN, is a code used to identify periodical publications, independent of which media are used (print and/or electronic). - Bioenergetics Communications, BEC: ISSN 2791-4690 |
| International System of Units | SI | The International System of Units (SI) is the modern form of the metric system of units for use in all aspects of life, including international trade, manufacturing, security, health and safety, protection of the environment, and in the basic science that underpins all of these. The system of quantities underlying the SI and the equations relating them are based on the present description of nature and are familiar to all scientists, technologists and engineers. The definition of the SI units is established in terms of a set of seven defining constants. The complete system of units can be derived from the fixed values of these defining constants, expressed in the units of the SI. These seven defining constants are the most fundamental feature of the definition of the entire system of units. These particular constants were chosen after having been identified as being the best choice, taking into account the previous definition of the SI, which was based on seven base units, and progress in science (p. 125). |
| International Union of Pure and Applied Chemistry, IUPAC | IUPAC | The International Union of Pure and Applied Chemistry (IUPAC) celebrated in 2019 the 100th anniversary, which coincided with the International Year of the Periodic Table of Chemical Elements (IYPT 2019). IUPAC {Quote} notes that marking Mendeleev's achievement will show how the periodic table is central to connecting cultural, economic, and political dimensions of global society “through a common language” {end of Quote} (Horton 2019). 2019 is proclaimed as the International Year of the Periodic Table of Chemical Elements (IYPT 2019). For a common language in mitochondrial physiology and bioenergetics, the IUPAC Green book (Cohen et al 2008) is a most valuable resource, which unfortunately is largely neglected in bioenergetics textbooks. Integration of open systems and non-equilibrium thermodynamic approaches remains a challenge for developing a common language (Gnaiger 1993; BEC 2020.1). |
| International oxygraph course | IOC | International Oxygraph Course (IOC), see O2k-Workshops. |
| ... further results | ||