Sadri 2023 Function (Oxf)
| Sadri S, Zhang X, Audi SH, Cowley AW Jr, Dash RK (2023) Computational modeling of substrate-dependent mitochondrial respiration and bioenergetics in the heart and kidney cortex and outer medulla. Function (Oxf) 4:zqad038. https://doi.org/10.1093/function/zqad038 |
Sadri S, Zhang X, Audi SH, Cowley AW Jr, Dash RK (2023) Function (Oxf)
Abstract: Integrated computational modeling provides a mechanistic and quantitative framework to characterize alterations in mitochondrial respiration and bioenergetics in response to different metabolic substrates in-silico. These alterations play critical roles in the pathogenesis of diseases affecting metabolically active organs such as heart and kidney. Therefore, the present study aimed to develop and validate thermodynamically constrained integrated computational models of mitochondrial respiration and bioenergetics in the heart and kidney cortex and outer medulla (OM). The models incorporated the kinetics of major biochemical reactions and transport processes as well as regulatory mechanisms in the mitochondria of these tissues. Intrinsic model parameters such as Michaelis–Menten constants were fixed at previously estimated values, while extrinsic model parameters such as maximal reaction and transport velocities were estimated separately for each tissue. This was achieved by fitting the model solutions to our recently published respirometry data measured in isolated rat heart and kidney cortex and OM mitochondria utilizing various NADH- and FADH2-linked metabolic substrates. The models were validated by predicting additional respirometry and bioenergetics data, which were not used for estimating the extrinsic model parameters. The models were able to predict tissue-specific and substrate-dependent mitochondrial emergent metabolic system properties such as redox states, enzyme and transporter fluxes, metabolite concentrations, membrane potential, and respiratory control index under diverse physiological and pathological conditions. The models were also able to quantitatively characterize differential regulations of NADH- and FADH2-linked metabolic pathways, which contribute differently toward regulations of oxidative phosphorylation and ATP synthesis in the heart and kidney cortex and OM mitochondria.
• Bioblast editor: Gnaiger E • O2k-Network Lab: US WI Milwaukee Dash RK
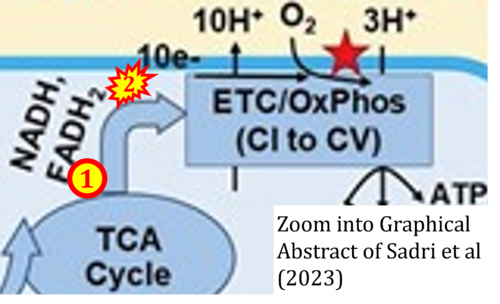
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
Labels: MiParea: Respiration
Organism: Rat
Tissue;cell: Heart, Kidney
Enzyme: Complex II;succinate dehydrogenase Regulation: Flux control Coupling state: OXPHOS Pathway: N, S, NS HRR: Oxygraph-2k