Difference between revisions of "Fatty acid oxidation pathway control state"
(Created page with "{{MitoPedia |description=right|300px|F-junction In the fatty acid oxidation or '''FAO'''-linked substrate state, one or several fatty acids are supplied...") |
|||
| Line 1: | Line 1: | ||
{{MitoPedia | {{MitoPedia | ||
|abbr=FAO, F | |||
|description=[[File:SUIT-Catg NF.jpg|right|300px|F-junction]] | |description=[[File:SUIT-Catg NF.jpg|right|300px|F-junction]] | ||
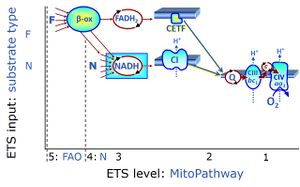
In the fatty acid oxidation or '''FAO'''-linked substrate state, one or several fatty acids are supplied to feed electrons into the [[F-junction]] through [[fatty acyl CoA dehydrogenase]] (reduced form [[FADH2]]), to [[electron transferring flavoprotein]] (CETF), and further through the [[Q-junction]] to [[Complex III]] (CIII). FAO not only depends on electron transfer through the F-junction (which is typically rate-limiting) but simultaneously generates NADH and thus depends on [[N-junction]] throughput. Hence FAO can be inhibited completely by inhibition of [[Complex I]] (CI). In addition and independent of this source of NADH, the type N substrate malate is required as a co-substrate for FAO in mt-preparations, since accumulation of AcetylCo inhibits FAO in the absence of malate. Malate is oxidized in a reaction catalyzed by malate dehydrogenase to oxaloacetate (yielding NADH), which then stimulates the entry of AcetylCo into the TCA cycle catalyzed by citrate synthase. | In the fatty acid oxidation or '''FAO'''-linked substrate state, one or several fatty acids are supplied to feed electrons into the [[F-junction]] through [[fatty acyl CoA dehydrogenase]] (reduced form [[FADH2]]), to [[electron transferring flavoprotein]] (CETF), and further through the [[Q-junction]] to [[Complex III]] (CIII). FAO not only depends on electron transfer through the F-junction (which is typically rate-limiting) but simultaneously generates NADH and thus depends on [[N-junction]] throughput. Hence FAO can be inhibited completely by inhibition of [[Complex I]] (CI). In addition and independent of this source of NADH, the type N substrate malate is required as a co-substrate for FAO in mt-preparations, since accumulation of AcetylCo inhibits FAO in the absence of malate. Malate is oxidized in a reaction catalyzed by malate dehydrogenase to oxaloacetate (yielding NADH), which then stimulates the entry of AcetylCo into the TCA cycle catalyzed by citrate synthase. | ||
|info=[[Gnaiger 2014 MitoPathways]] | |info=[[Gnaiger 2014 MitoPathways]] | ||
}} | }} | ||
{{MitoPedia methods | {{MitoPedia concepts}} | ||
{{MitoPedia methods}} | |||
}} | |||
{{MitoPedia O2k and high-resolution respirometry | {{MitoPedia O2k and high-resolution respirometry | ||
|mitopedia O2k and high-resolution respirometry=SUIT | |mitopedia O2k and high-resolution respirometry=SUIT state | ||
}} | }} | ||
{{MitoPedia topics}} | |||
Revision as of 20:01, 9 May 2016
- high-resolution terminology - matching measurements at high-resolution
Fatty acid oxidation pathway control state
Description
In the fatty acid oxidation or FAO-linked substrate state, one or several fatty acids are supplied to feed electrons into the F-junction through fatty acyl CoA dehydrogenase (reduced form FADH2), to electron transferring flavoprotein (CETF), and further through the Q-junction to Complex III (CIII). FAO not only depends on electron transfer through the F-junction (which is typically rate-limiting) but simultaneously generates NADH and thus depends on N-junction throughput. Hence FAO can be inhibited completely by inhibition of Complex I (CI). In addition and independent of this source of NADH, the type N substrate malate is required as a co-substrate for FAO in mt-preparations, since accumulation of AcetylCo inhibits FAO in the absence of malate. Malate is oxidized in a reaction catalyzed by malate dehydrogenase to oxaloacetate (yielding NADH), which then stimulates the entry of AcetylCo into the TCA cycle catalyzed by citrate synthase.
Abbreviation: FAO, F
Reference: Gnaiger 2014 MitoPathways
MitoPedia O2k and high-resolution respirometry: "SUIT state" is not in the list (O2k hardware, DatLab, Oroboros QM, O2k-Open Support, O2k-Respirometry, O2k-FluoRespirometry) of allowed values for the "MitoPedia O2k and high-resolution respirometry" property.
SUIT state"SUIT state" is not in the list (O2k hardware, DatLab, Oroboros QM, O2k-Open Support, O2k-Respirometry, O2k-FluoRespirometry) of allowed values for the "MitoPedia O2k and high-resolution respirometry" property.