Difference between revisions of "Enable DL-Protocol editing"
Tindle Lisa (talk | contribs) |
Tindle Lisa (talk | contribs) m |
||
| (6 intermediate revisions by 2 users not shown) | |||
| Line 2: | Line 2: | ||
__TOC__ | __TOC__ | ||
{{MitoPedia without banner | {{MitoPedia without banner | ||
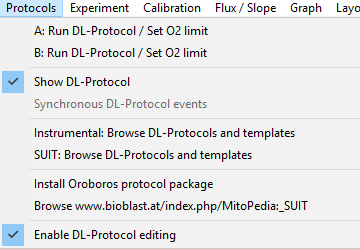
|description= | |description= '''Enable DL-Protocol editing''' is a novel function of DatLab 7.4 offering a new feature in DL-Protocols: flexibility. Fixed sequences of events and marks can be changed (Skip/Added) in a SUIT protocol by the user. Moreover, the text, instructions, concentrations and titration volumes of injections in a specific DL-Protocol can be edited and saved as [[Export_DL-Protocol_User_(*.DLPU)| user-specific DL-Protocol]] [File]\Export\DL-Protocol User (*.DLPU). To enable it, under the 'Protocols' tab in the menu, select the option 'Enable DL-Protocol editing', and then select the plot in which the marks will be set (''e.g.,'' O2 flux per V). Select the 'Overview' window, where you will be able to edit events and marks names, definition/state, final concentration and titration volumes, as well as select a mark as 'multi' for multiple titration steps, skip a mark, or add a new event or mark. After saving, [[Export_DL-Protocol_User_(*.DLPU)|export a DL-Protocol User (DLPU)]] and load it before running the next experiments. If users of DatLab versions older than DatLab 7.4 wish to alter the nature of the chemicals used or the sequence of injections, we ask them to [https://www.oroboros.at/index.php/o2k-technical-support/ contact the O2k-Technical Support]. | ||
For more information: | For more information: | ||
Latest revision as of 13:05, 26 July 2021
 |
Enable DL-Protocol editing |
MitoPedia O2k and high-resolution respirometry:
O2k-Open Support
Description
Enable DL-Protocol editing is a novel function of DatLab 7.4 offering a new feature in DL-Protocols: flexibility. Fixed sequences of events and marks can be changed (Skip/Added) in a SUIT protocol by the user. Moreover, the text, instructions, concentrations and titration volumes of injections in a specific DL-Protocol can be edited and saved as user-specific DL-Protocol [File]\Export\DL-Protocol User (*.DLPU). To enable it, under the 'Protocols' tab in the menu, select the option 'Enable DL-Protocol editing', and then select the plot in which the marks will be set (e.g., O2 flux per V). Select the 'Overview' window, where you will be able to edit events and marks names, definition/state, final concentration and titration volumes, as well as select a mark as 'multi' for multiple titration steps, skip a mark, or add a new event or mark. After saving, export a DL-Protocol User (DLPU) and load it before running the next experiments. If users of DatLab versions older than DatLab 7.4 wish to alter the nature of the chemicals used or the sequence of injections, we ask them to contact the O2k-Technical Support.
For more information:
 Export DL-Protocol User (*.DLPU)
Export DL-Protocol User (*.DLPU)
MitoPedia methods:
Respirometry
MitoPedia O2k and high-resolution respirometry:
DatLab