Difference between revisions of "OctPGM pathway control state"
From Bioblast
| (13 intermediate revisions by 5 users not shown) | |||
| Line 3: | Line 3: | ||
|description='''OctPGM''': [[Octanoylcarnitine]] & [[Pyruvate]] & [[Glutamate]] & [[Malate]]. | |description='''OctPGM''': [[Octanoylcarnitine]] & [[Pyruvate]] & [[Glutamate]] & [[Malate]]. | ||
'''MitoPathway control:''' | '''MitoPathway control state:''' [[FN]] | ||
'''SUIT protocols:''' [[ | '''SUIT protocols:''' [[SUIT-002]] | ||
|info=[[MiPNet21.06 SUIT | :This substrate combination supports N-linked flux which is typically higher than FAO capacity (F/FN<1 in the OXPHOS state). In SUIT-RP1, PMOct is induced after PM(E), to evaluate any additive effect of adding Oct. In SUIT-RP2, FAO OXPHOS capacity is measured first, testing for the effect of increasing malate concentration (compare [[malate-anaplerotic pathway control state]], M alone), and pyruvate and glutamate is added to compare FAO as the background state with FN as the reference state. | ||
|info=[[MiPNet21.06 SUIT RP]] | |||
}} | }} | ||
::: ''More details'' | |||
| | ::::» [[FN]] | ||
::::» [[Additive effect of convergent electron flow]] | |||
::::» [[Respiratory_complexes#Respiratory_complexes_-_more_than_five |Respiratory complexes - more than five]] | |||
::::» [[OctGM]] | |||
::::» [[Convergent electron flow]] | |||
== OctPGM<sub>''L''</sub> == | |||
FN pathway in the LEAK state can be evaluated in the following SUIT protocols: | |||
== OctPGM | == OctPGM<sub>''P''</sub> == | ||
FN pathway in the OXPHOS state can be evaluated in the following SUIT protocols: | |||
:::*[[SUIT-002]] | |||
::::* DL-Protocol for isolated mitochondria and tissue homogenate (mt): [[SUIT-002 O2 mt D005]] | |||
::::* DL-Protocol for permeabilized fibers (pfi): [[SUIT-002 O2 pfi D006]] | |||
::::* DL-Protocol for permeabilized cells (pce): [[SUIT-002 O2 ce-pce D007]] | |||
:::*[[SUIT-015]] | |||
== OctPGM<sub>''E''</sub> == | |||
FN pathway in the ET state can be evaluated in the following SUIT protocols: | |||
---- | ---- | ||
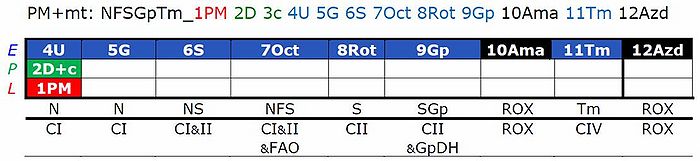
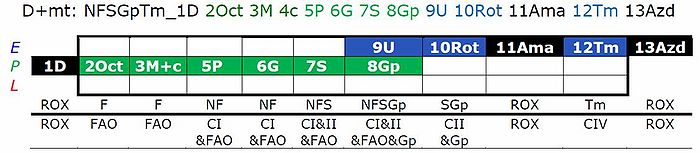
[[File:SUIT-Reference-Protocol 1.jpg|700px | | [[File:SUIT-Reference-Protocol 1.jpg|700px |1PM;2D;3U;4G;5S;6Oct;7Rot;8Gp-]]<br /> | ||
| Line 32: | Line 42: | ||
---- | ---- | ||
{{MitoPedia concepts | |||
|mitopedia concept=SUIT state | |||
}} | |||
Latest revision as of 05:00, 19 July 2022
Description
OctPGM: Octanoylcarnitine & Pyruvate & Glutamate & Malate.
MitoPathway control state: FN
SUIT protocols: SUIT-002
- This substrate combination supports N-linked flux which is typically higher than FAO capacity (F/FN<1 in the OXPHOS state). In SUIT-RP1, PMOct is induced after PM(E), to evaluate any additive effect of adding Oct. In SUIT-RP2, FAO OXPHOS capacity is measured first, testing for the effect of increasing malate concentration (compare malate-anaplerotic pathway control state, M alone), and pyruvate and glutamate is added to compare FAO as the background state with FN as the reference state.
Abbreviation: OctPGM
Reference: MiPNet21.06 SUIT RP
OctPGML
FN pathway in the LEAK state can be evaluated in the following SUIT protocols:
OctPGMP
FN pathway in the OXPHOS state can be evaluated in the following SUIT protocols:
-
- DL-Protocol for isolated mitochondria and tissue homogenate (mt): SUIT-002 O2 mt D005
- DL-Protocol for permeabilized fibers (pfi): SUIT-002 O2 pfi D006
- DL-Protocol for permeabilized cells (pce): SUIT-002 O2 ce-pce D007
-
OctPGME
FN pathway in the ET state can be evaluated in the following SUIT protocols:
MitoPedia concepts:
SUIT state