Difference between revisions of "Run DL-Protocol/Set O2 limit"
| Line 16: | Line 16: | ||
<div id="DL-Protocols-anchor4"></div> | <div id="DL-Protocols-anchor4"></div> | ||
::::'''Synchronous DL-Protocol events'''. This option is only available when the same DL-Protocol is selected (in [[#DL-Protocols-anchor1|'Set DL-Protocol / O2 limit']]) for chamber A and B (protocol names are displayed in the name field of the [[DL-Protocols#DL-Protocol window|DL-Protocol window]]). When checked [[File:Check-mark.png|17px]], the user only needs to follow one protocol sequence of either chamber A or B. For example, when an event is set for chamber A, it will also be automatically set for chamber B - the 'Event Information' window indicates "Set event in both chambers" in bold letters. | ::::'''Synchronous DL-Protocol events'''. This option is only available when the same DL-Protocol is selected (in [[#DL-Protocols-anchor1|'Set DL-Protocol / O2 limit']]) for chamber A and B (protocol names are displayed in the name field of the [[DL-Protocols#DL-Protocol window|DL-Protocol window]]). When checked [[File:Check-mark.png|17px]], the user only needs to follow one protocol sequence of either chamber A or B. For example, when an event is set for chamber A, it will also be automatically set for chamber B - the 'Event Information' window indicates "Set event in both chambers" in bold letters. | ||
[[File:Set_DL-_Protocol_O2_-_window.png|thumb|right|200px|'''Set DL-Protocol / O2 limit''' For chamber A (upper frame) a specific protocol is selected, showing the name ("RP1mt"), the hyperlink to the | [[File:Set_DL-_Protocol_O2_-_window.png|thumb|right|200px|'''Set DL-Protocol / O2 limit''' For chamber A (upper frame) a specific protocol is selected, showing the name ("RP1mt"), the hyperlink to the Biobast in blue and additional information. Furthermore, the 'Lower O2 limit [µM]' alert is checked. In chamber B (lower frame) no DL-protocol is selected.]] | ||
| Line 22: | Line 22: | ||
::::'''Set DL-Protocol / O2 limit'''. In the 'Set DL-Protocol / O2 limit' window a DL-Protocol can be assigned to specific chamber. | ::::'''Set DL-Protocol / O2 limit'''. In the 'Set DL-Protocol / O2 limit' window a DL-Protocol can be assigned to specific chamber. | ||
:::::*'''Buttons'''. DL-protocols (either a .DLP or .DLPU file) can be selected ([[File:Select-button.png|50px]]) for chamber A and B. '.DLP' is the original file format for DL-Protocols provided by | :::::*'''Buttons'''. DL-protocols (either a .DLP or .DLPU file) can be selected ([[File:Select-button.png|50px]]) for chamber A and B. '.DLP' is the original file format for DL-Protocols provided by Oroboros, '.DLPU' files are user-modified versions (see [[#DL-Protocols-anchor3|A: Export DL-Protocol]] and [[#DL-Protocols-anchor2|'Edit DL-Protocol' window]]) of a specific DL-Protocols ('.DLP' file). Only when a DL-Protocol is initially selected, the [[File:Show_this_file-button.png|60px]] button ist active and enables to automatically open the DL-Protocol file in another DatLab instance to see a DL-Protocol specific example data plot with properly set marks and events. Clicking the [[File:Clear-button.png|50px]] button removes the selected DL-Protocol. | ||
:::::*'''Additional information'''. When a DL-protocol is selected for chamber A or B further text information is provided in the chamber specific field: (1) protocol name, (2) a hyperlink (in blue, underlined) to the protocol in the Bioblast and (3) additional information and short description for the DL-Protocol. | :::::*'''Additional information'''. When a DL-protocol is selected for chamber A or B further text information is provided in the chamber specific field: (1) protocol name, (2) a hyperlink (in blue, underlined) to the protocol in the Bioblast and (3) additional information and short description for the DL-Protocol. | ||
[[File:Warning-O2.png|thumb|right|200px|'''O2-limit warning button''']] | [[File:Warning-O2.png|thumb|right|200px|'''O2-limit warning button''']] | ||
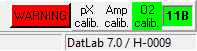
:::::*'''Lower O2 limit [µM]'''. When checked, an automatic warning notifies the user when the actual | :::::*'''Lower O2 limit [µM]'''. When checked, an automatic warning notifies the user when the actual oxygen level falls below the specified value for the according chamber: A red flashing "WARNING" button in the status line (DatLab window bottom, right) appears and a "WARNING" event is set in the plot. To reset the actual warning status (to notify the next possible fall-below), the red flashing "WARNING" button has to be clicked. First, a 'Warning window' opens to show all warnings during the experiment. After closing the window, the reset is completed. | ||
| Line 38: | Line 38: | ||
== DL-Protocol window == | == DL-Protocol window == | ||
[[File:DL-Protocol window.png|thumb|right|200px|'''DL-Protocol window''' in DatLab, highlighted by a red box in the image. The DL-Protocol windows for chamber A (up) and chamber B (down) are shown with different DL-protocols as indicated by the DL-protocol name in the name field (1, red oval)]] | [[File:DL-Protocol window.png|thumb|right|200px|'''DL-Protocol window''' in DatLab, highlighted by a red box in the image. The DL-Protocol windows for chamber A (up) and chamber B (down) are shown with different DL-protocols as indicated by the DL-protocol name in the name field (1, red oval)]] | ||
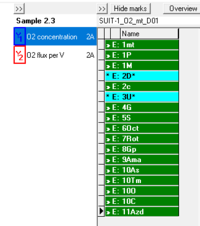
::::'''The DL-Protocol window''' appears to the very right for every chamber. It | ::::'''The DL-Protocol window''' appears to the very right for every chamber. It consists of three buttons at the top, a name field and a list to display events and marks. | ||
::::*'''Buttons'''. The [[File:_Button_show-hide_DL-Protocol_window.png|17px]] button is used to hide/show the DL-Protocol window (similar to 'Show DL-Protocol' in the 'Protocol' menu). Clicking the [[File:Hide_marks.png|x17px]]/[[File:Show_marks.png|x17px]] button toggles between hiding or showing the marks in the list. When the marks in the DL-Protocol are hidden the user only needs to focus on the sequence of events, facilitating the experimental procedure.The [[File:Edit.png|60px]] button opens the [[#DL-Protocols-anchor2|'Edit DL-Protocol' window]] to make changes in events and marks in the DL-Protocol. | ::::*'''Buttons'''. The [[File:_Button_show-hide_DL-Protocol_window.png|17px]] button is used to hide/show the DL-Protocol window (similar to 'Show DL-Protocol' in the 'Protocol' menu). Clicking the [[File:Hide_marks.png|x17px]]/[[File:Show_marks.png|x17px]] button toggles between hiding or showing the marks in the list. When the marks in the DL-Protocol are hidden the user only needs to focus on the sequence of events, facilitating the experimental procedure. The [[File:Edit.png|60px]] button opens the [[#DL-Protocols-anchor2|'Edit DL-Protocol' window]] to make changes in events and marks in the DL-Protocol. | ||
::::*'''Information'''. Below the buttons, the 'name field' automatically displays the name of the loaded protocol. | ::::*'''Information'''. Below the buttons, the 'name field' automatically displays the name of the loaded protocol. | ||
| Line 48: | Line 48: | ||
[[File:Edit_DL-protocol_window.png|thumb|right|200px|alt=Alt text|link=[[Media:Edit_DL-protocol_window.png]]|'''Edit DL-protocol window''' ...]] | [[File:Edit_DL-protocol_window.png|thumb|right|200px|alt=Alt text|link=[[Media:Edit_DL-protocol_window.png]]|'''Edit DL-protocol window''' ...]] | ||
<div id="DL-Protocols-anchor2"></div> | <div id="DL-Protocols-anchor2"></div> | ||
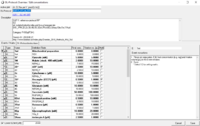
::::''''Edit DL-Protocol' window'''. In the listed events and marks, changes can be made in | ::::''''Edit DL-Protocol' window'''. In the listed events and marks, changes can be made in 'Final conc. [mM]', 'Titration vol. [µL]' and 'Multi' but not in 'Type', 'Name' and 'Definition/State'. Further changes are accepted in 'Comment' and 'DL-Protocol description' at the bottom of the window. | ||
:::::*'''Information''' in the window head. The active plot is indicated in the according name field (window, top). A second name field displays the DL-Protocol name, furthermore, a hyperlink (blue, underlined) to the | :::::*'''Information''' in the window head. The active plot is indicated in the according name field (window, top). A second name field displays the DL-Protocol name, furthermore, a hyperlink (blue, underlined) to the Biobast is available. | ||
<div id="DL-Protocols-anchor5"></div> | <div id="DL-Protocols-anchor5"></div> | ||
| Line 55: | Line 55: | ||
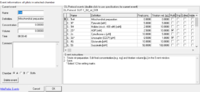
:::::**'''Type''' and '''Name'''. Similar to the [[DL-Protocols#DL-Protocol window|DL-Protocol window]], mark names are preceded by "'''>> M:'''", events are indicated by "'''>> E:'''" or "'''<big>∗</big> E:'''" (multi-events). | :::::**'''Type''' and '''Name'''. Similar to the [[DL-Protocols#DL-Protocol window|DL-Protocol window]], mark names are preceded by "'''>> M:'''", events are indicated by "'''>> E:'''" or "'''<big>∗</big> E:'''" (multi-events). | ||
:::::**'''Final conc. [mM]''' and '''Titration vol. [µL]'''. The fields contain the according values for an event or mark and can be changed by the user. | :::::**'''Final conc. [mM]''' and '''Titration vol. [µL]'''. The fields contain the according values for an event or mark and can be changed by the user. | ||
:::::**'''Multi'''. To toggle an events status from or to the status 'multi-event' ,the checkbox in the column has to be set [[File:List-ckeckbox-checked.png|15px]] or unchecked [[File:List-ckeckbox-unchecked.png|15px]], | :::::**'''Multi'''. To toggle an events status from or to the status 'multi-event' ,the checkbox in the column has to be set [[File:List-ckeckbox-checked.png|15px]] or unchecked [[File:List-ckeckbox-unchecked.png|15px]], whereupon the indicator ("'''>>'''" or "'''<big>∗</big> '''") for the event changes. For marks, a set checkbox is of no relevance. | ||
:::::*'''Comment'''. This field contains specific information for an event or mark and can be changed/edited by the user. | :::::*'''Comment'''. This field contains specific information for an event or mark and can be changed/edited by the user. | ||
| Line 68: | Line 68: | ||
::::*'''Events and marks basics'''. | ::::*'''Events and marks basics'''. | ||
::::**The [[#DL-Protocol window|'DL-Protocol window']] shows the | ::::**The [[#DL-Protocol window|'DL-Protocol window']] shows the sequence of '''DL-Protocol events''' and '''DL-Protocol marks''' that have to be set to complete an experiment. Besides, it is possible to set '''user-defined marks''' and '''user-defined events''' (menu 'Experiment' -> 'Add event' or F4 or STRG-left-click in plot), which will not affect the DL-Protocol and will be stored together with the DL-Protocol marks and events in the .DLD data file. Extra '''user-defined events''' are of importance for unpredictable occurrences e.g. when a chamber has to be re-oxygenised, requiring 'open' and 'close' events. | ||
::::**It is possible to edit/change a '''user-defined mark''' or '''user-defined event''' into '''DL-Protocol mark''' or '''DL-Protocol event''', respectively. | ::::**It is possible to edit/change a '''user-defined mark''' or '''user-defined event''' into '''DL-Protocol mark''' or '''DL-Protocol event''', respectively. | ||
::::**In most instances, a '''DL-Protocol event''' is a titration step of a specific substance according to the DL Protocol. Thereby, the DL-Protocol event demarcates a specific 'state' (in particular a [[MitoPedia:_Respiratory_states|respiratory state]], depending on the titrated substance) until the next DL-Protocol event. Within a 'states' a DL-Protocol mark is set to read out the state-specific [[Oxygen_flux|oxygen flux]] (or any other signal such as oxygen or fluorescence - has to be defined in the DL-Protocol). | ::::**In most instances, a '''DL-Protocol event''' is a titration step of a specific substance according to the DL Protocol. Thereby, the DL-Protocol event demarcates a specific 'state' (in particular a [[MitoPedia:_Respiratory_states|respiratory state]], depending on the titrated substance) until the next DL-Protocol event. Within a 'states' a DL-Protocol mark is set to read out the state-specific [[Oxygen_flux|oxygen flux]] (or any other signal such as oxygen or fluorescence - has to be defined in the DL-Protocol). | ||
| Line 76: | Line 76: | ||
::::*'''Events in action'''. An HRFR experiment is primarily an '''event'''-driven process: '''DL-Protocol events''' MUST be set; '''DL-Protocol marks''' DON'T NEED to be regarded at first, when running the DL-Protocol. Hence, for practicability, the '''DL-Protocol marks''' can be hidden in the [[#DL-Protocols-anchor5|list]] ([[#DL-Protocol window|'DL-Protocol window']]: [[File:Hide_marks.png|60px]]) and the user only has to focus on the sequence of the '''DL-Protocol events'''. Still, '''DL-Protocol marks''' or any '''user-defined marks''' can be set at any time during the experimental run. | ::::*'''Events in action'''. An HRFR experiment is primarily an '''event'''-driven process: '''DL-Protocol events''' MUST be set; '''DL-Protocol marks''' DON'T NEED to be regarded at first, when running the DL-Protocol. Hence, for practicability, the '''DL-Protocol marks''' can be hidden in the [[#DL-Protocols-anchor5|list]] ([[#DL-Protocol window|'DL-Protocol window']]: [[File:Hide_marks.png|60px]]) and the user only has to focus on the sequence of the '''DL-Protocol events'''. Still, '''DL-Protocol marks''' or any '''user-defined marks''' can be set at any time during the experimental run. | ||
::::*'''Marks in action''' For practicability, it is recommended to set the '''DL-Protocol marks''' after the experimental procedure has been completed. By definition, a DL-Protocol mark is necessarily linked to a | ::::*'''Marks in action''' For practicability, it is recommended to set the '''DL-Protocol marks''' after the experimental procedure has been completed. By definition, a DL-Protocol mark is necessarily linked to a preceding DL-Protocol event, as determined in DL-Protocol sequence. Only when set behind the corresponding DL-Protocol event, The DL-Protocol mark is correctly set and the name is automatically assigned. With DL-Protocol events set in a plot, the DL-Protocol marks can be set in any order, because the automatic mark naming is based on the preceding DL-Protocol event. When setting a '''second or any further''' mark in a state (behind a DL-Protocol event), this mark will be recognised as a '''user-defined mark''', automatically named with a single letter (alphabetically, starting with 'a'). The same is true, when a mark is set in the plot behind an DL-Protocol event, where no DL-Protocol mark is defined in the DL-Protocol. | ||
| Line 108: | Line 108: | ||
| mark | | mark | ||
| "'''>> M:'''" | | "'''>> M:'''" | ||
| The DL-Protocol mark | | The DL-Protocol mark occurs at the right position, after the associated DL-Protocol event. | ||
|- | |- | ||
|rowspan="2" style="background:#FF0000"| Red | |rowspan="2" style="background:#FF0000"| Red | ||
| events | | events | ||
| "'''>> E:'''" or "'''<big>∗</big> E:'''" | | "'''>> E:'''" or "'''<big>∗</big> E:'''" | ||
| (i) The DL-Protocol event occurs in the wrong position e.g. a later on DL-Protocol event from the DL-Protocol | | (i) The DL-Protocol event occurs in the wrong position e.g. a later on DL-Protocol event from the DL-Protocol sequence has been set too early. (ii) A scheduled DL-Protocol event is missing in(has been left out) or has been deleted from the processed sequence - can lead to a sequence of red events. (iii) The ""'''>> E:'''" DL-Protocol event (not a multi-event!) occurs multiple times in the plot. | ||
|- | |- | ||
| marks | | marks | ||
| Line 134: | Line 134: | ||
::::*'''Start'''. Hardware and Software must be setup properly (refer to the [[MiPNet22.11_O2k-FluoRespirometer_manual|O2k-FluoRespirometer manual]]). DatLab 7 has to be started, connected ('Connect to O2k') and all instrumental- and experimental parameters have to be set. | ::::*'''Start'''. Hardware and Software must be setup properly (refer to the [[MiPNet22.11_O2k-FluoRespirometer_manual|O2k-FluoRespirometer manual]]). DatLab 7 has to be started, connected ('Connect to O2k') and all instrumental- and experimental parameters have to be set. | ||
::::*'''Loading a DL-Protocol'''. A DL-Protocol (either .DLP or DLPU) can be loaded for each chamber in [[#DL-Protocols-anchor1|'Set DL-Protocol / O2 limit']]. When the same DL-Protocol is loaded for both chambers, the [[#DL-Protocols-anchor4|'Synchronous DL-Protocol events']] mode of operation is available. The loaded DL-protocols for each chamber are | ::::*'''Loading a DL-Protocol'''. A DL-Protocol (either .DLP or DLPU) can be loaded for each chamber in [[#DL-Protocols-anchor1|'Set DL-Protocol / O2 limit']]. When the same DL-Protocol is loaded for both chambers, the [[#DL-Protocols-anchor4|'Synchronous DL-Protocol events']] mode of operation is available. The loaded DL-protocols for each chamber are displayed in the [[#DL-Protocol window|'DL-Protocol window']] to the right in DatLab 7. | ||
[[File:Event_info_SET_chamber_A.png|thumb|right|200px|'''Event information window - Set event''')]] | [[File:Event_info_SET_chamber_A.png|thumb|right|200px|'''Event information window - Set event''')]] | ||
::::*'''Running a DL-Protocol'''. Once the DL-Protocol is loaded, the protocol layout will be displayed on the right side in the [[#DL-Protocol window|'DL-Protocol window']]. The experiment can now be started following the '''DL Protocol event''' sequence. For this purpose, left-klick onto the first '''DL Protocol event''' in yellow (<span style="background-color:#FFFF00; border-color:#D4D0D0; border-style: solid; border-width: 1px; margin: 2px">'''>> E: xxx'''</span>) of the DL Protocol and the Event information window will open to set the DL-Protocol event in the plot. When the DL-Protocol event is set correctly according to the protocol sequence it turns into green (<span style="background-color:#377A34; border-color:#D4D0D0; border-style: solid; border-width: 1px; margin: 2px; color:#FFFFFF">'''>> E: xxx'''</span>) in the DL-Protocol window list. | ::::*'''Running a DL-Protocol'''. Once the DL-Protocol is loaded, the protocol layout will be displayed on the right side in the [[#DL-Protocol window|'DL-Protocol window']]. The experiment can now be started following the '''DL Protocol event''' sequence. For this purpose, left-klick onto the first '''DL Protocol event''' in yellow (<span style="background-color:#FFFF00; border-color:#D4D0D0; border-style: solid; border-width: 1px; margin: 2px">'''>> E: xxx'''</span>) of the DL Protocol and the Event information window will open to set the DL-Protocol event in the plot. When the DL-Protocol event is set correctly according to the protocol sequence it turns into green (<span style="background-color:#377A34; border-color:#D4D0D0; border-style: solid; border-width: 1px; margin: 2px; color:#FFFFFF">'''>> E: xxx'''</span>) in the DL-Protocol window list. Proceed with the next yellow DL-Protocol event <span style="background-color:#FFFF00; border-color:#D4D0D0; border-style: solid; border-width: 1px; margin: 2px">'''>> E: yyy'''</span> at the according time point (after flux has stabilized or experiment specific timing). When a multi-event is encountered the first time to be set it appears in yellow (<span style="background-color:#FFFF00; border-color:#D4D0D0; border-style: solid; border-width: 1px; margin: 2px">'''<big>∗</big> E: xxx <big>*</big>'''</span>). After that it appears in auqua (<span style="background-color:#00FFFF; border-color:#D4D0D0; border-style: solid; border-width: 1px; margin: 2px">'''<big>∗</big> E: xxx <big>*</big>'''</span>) and can be set multiple times, according to the experimental needs. | ||
<!-- | |||
<!-- | |||
::::DL-Protocol will inform you if a titration event is missed or a repetition of a non multiple titration occured (highlighted in red).--> | ::::DL-Protocol will inform you if a titration event is missed or a repetition of a non multiple titration occured (highlighted in red).--> | ||
Revision as of 15:24, 28 July 2017
Description
DL-Protocols can be selected in DatLab 7 in the pull-down menu 'Protocol': Set DL-Protocol / O2 limit. A DL-Protocol defines the sequence of Events and Marks. Instrumental DL-Protocols are used for calibrations and instrumental quality control, typically without experimental sample in the incubation medium. DL-Protocols for substrate-uncoupler-inhibitor titrations (see MitoPedia: SUIT) proceed stepwise to activate a sequence of coupling control states and pathway control states. A specific SUIT protocol can be assigned to O2k-chamber A or B or both. The Titration-Injection-microPump TIP2k can be programmed for automatic control of titration steps in a DL-Protocol. A collection of evaluated and tested standard DL-Protocols are provided. These can be edited and saved under 'Lab protocols' or under the User logged into DatLab. A Lower O2 limit [µM] can be defined for each chamber, to trigger an automatic warning when the experimental O2 concentration declines below this limit as a WARNING to remind the user that re-oxygenation of the chamber is required.
Abbreviation: DLP
MitoPedia O2k and high-resolution respirometry:
DatLab
- Show DL-Protocol can be check-marked (
 ) to hide or show the DL-Protocol window. Alternatively, the
) to hide or show the DL-Protocol window. Alternatively, the  button (upper left) in the DL-Protocol window can be used to hide or show the window. When DatLab is started for data recording, an empty DL-Protocol window shows up because no DL-protocol is specified at this moment.
button (upper left) in the DL-Protocol window can be used to hide or show the window. When DatLab is started for data recording, an empty DL-Protocol window shows up because no DL-protocol is specified at this moment.
- Show DL-Protocol can be check-marked (
- Synchronous DL-Protocol events. This option is only available when the same DL-Protocol is selected (in 'Set DL-Protocol / O2 limit') for chamber A and B (protocol names are displayed in the name field of the DL-Protocol window). When checked
 , the user only needs to follow one protocol sequence of either chamber A or B. For example, when an event is set for chamber A, it will also be automatically set for chamber B - the 'Event Information' window indicates "Set event in both chambers" in bold letters.
, the user only needs to follow one protocol sequence of either chamber A or B. For example, when an event is set for chamber A, it will also be automatically set for chamber B - the 'Event Information' window indicates "Set event in both chambers" in bold letters.
- Synchronous DL-Protocol events. This option is only available when the same DL-Protocol is selected (in 'Set DL-Protocol / O2 limit') for chamber A and B (protocol names are displayed in the name field of the DL-Protocol window). When checked
- Set DL-Protocol / O2 limit. In the 'Set DL-Protocol / O2 limit' window a DL-Protocol can be assigned to specific chamber.
- Buttons. DL-protocols (either a .DLP or .DLPU file) can be selected (
 ) for chamber A and B. '.DLP' is the original file format for DL-Protocols provided by Oroboros, '.DLPU' files are user-modified versions (see A: Export DL-Protocol and 'Edit DL-Protocol' window) of a specific DL-Protocols ('.DLP' file). Only when a DL-Protocol is initially selected, the
) for chamber A and B. '.DLP' is the original file format for DL-Protocols provided by Oroboros, '.DLPU' files are user-modified versions (see A: Export DL-Protocol and 'Edit DL-Protocol' window) of a specific DL-Protocols ('.DLP' file). Only when a DL-Protocol is initially selected, the  button ist active and enables to automatically open the DL-Protocol file in another DatLab instance to see a DL-Protocol specific example data plot with properly set marks and events. Clicking the
button ist active and enables to automatically open the DL-Protocol file in another DatLab instance to see a DL-Protocol specific example data plot with properly set marks and events. Clicking the  button removes the selected DL-Protocol.
button removes the selected DL-Protocol.
- Buttons. DL-protocols (either a .DLP or .DLPU file) can be selected (
- Additional information. When a DL-protocol is selected for chamber A or B further text information is provided in the chamber specific field: (1) protocol name, (2) a hyperlink (in blue, underlined) to the protocol in the Bioblast and (3) additional information and short description for the DL-Protocol.
- Lower O2 limit [µM]. When checked, an automatic warning notifies the user when the actual oxygen level falls below the specified value for the according chamber: A red flashing "WARNING" button in the status line (DatLab window bottom, right) appears and a "WARNING" event is set in the plot. To reset the actual warning status (to notify the next possible fall-below), the red flashing "WARNING" button has to be clicked. First, a 'Warning window' opens to show all warnings during the experiment. After closing the window, the reset is completed.
- A: Export DL-Protocol. The user can save changes in a DL-Protocol and export an user-modified DL-Protocol (.DLPU) for reuse. To make changes in a DL-Protocol see: 'Edit DL-Protocol' window
- B: Export DL-Protocol. applies for chamber B as described above in 'A: Export DL-Protocol'.
DL-Protocol window
- The DL-Protocol window appears to the very right for every chamber. It consists of three buttons at the top, a name field and a list to display events and marks.
- Buttons. The
 button is used to hide/show the DL-Protocol window (similar to 'Show DL-Protocol' in the 'Protocol' menu). Clicking the
button is used to hide/show the DL-Protocol window (similar to 'Show DL-Protocol' in the 'Protocol' menu). Clicking the  /
/ button toggles between hiding or showing the marks in the list. When the marks in the DL-Protocol are hidden the user only needs to focus on the sequence of events, facilitating the experimental procedure. The
button toggles between hiding or showing the marks in the list. When the marks in the DL-Protocol are hidden the user only needs to focus on the sequence of events, facilitating the experimental procedure. The  button opens the 'Edit DL-Protocol' window to make changes in events and marks in the DL-Protocol.
button opens the 'Edit DL-Protocol' window to make changes in events and marks in the DL-Protocol.
- Buttons. The
- Information. Below the buttons, the 'name field' automatically displays the name of the loaded protocol.
- List. The list in the DL-Protocol window shows the sequence of events and marks. The numbers in the event and mark names corresponds to the defined sequence in the DL-Protocol. Mark names are preceded by ">> M:", events are indicated by ">> E:" or "∗ E:". Every mark or event is unique and must be set only once. However, there is an exception for events identified by "∗", which can be set consecutively multiple times (multi-events), as it is necessary for titrating the uncoupler (U) in SUIT protocols. The color-code for DL-Protocol marks and events in the list are described below in 'DL-Protocol colors'.
- 'Edit DL-Protocol' window. In the listed events and marks, changes can be made in 'Final conc. [mM]', 'Titration vol. [µL]' and 'Multi' but not in 'Type', 'Name' and 'Definition/State'. Further changes are accepted in 'Comment' and 'DL-Protocol description' at the bottom of the window.
- Information in the window head. The active plot is indicated in the according name field (window, top). A second name field displays the DL-Protocol name, furthermore, a hyperlink (blue, underlined) to the Biobast is available.
- 'Edit DL-Protocol' window. In the listed events and marks, changes can be made in 'Final conc. [mM]', 'Titration vol. [µL]' and 'Multi' but not in 'Type', 'Name' and 'Definition/State'. Further changes are accepted in 'Comment' and 'DL-Protocol description' at the bottom of the window.
- List. In the list of the window, events are always displayed, however, only the marks (if available) for the active plot (O2 or O2 flux) are shown.
- Type and Name. Similar to the DL-Protocol window, mark names are preceded by ">> M:", events are indicated by ">> E:" or "∗ E:" (multi-events).
- Final conc. [mM] and Titration vol. [µL]. The fields contain the according values for an event or mark and can be changed by the user.
- Multi. To toggle an events status from or to the status 'multi-event' ,the checkbox in the column has to be set
 or unchecked
or unchecked  , whereupon the indicator (">>" or "∗ ") for the event changes. For marks, a set checkbox is of no relevance.
, whereupon the indicator (">>" or "∗ ") for the event changes. For marks, a set checkbox is of no relevance.
- List. In the list of the window, events are always displayed, however, only the marks (if available) for the active plot (O2 or O2 flux) are shown.
- Comment. This field contains specific information for an event or mark and can be changed/edited by the user.
- DL-Protocol description. This field contains general, DL-Protocol specific information and can be changed/edited by the user.
- Changes can be accepted
 or rejected
or rejected  . Finally, changes in the DL-Protocol can be saved/exported to generate a user modified DL-Protocol (.DLPU), see A: Export DL-Protocol.
. Finally, changes in the DL-Protocol can be saved/exported to generate a user modified DL-Protocol (.DLPU), see A: Export DL-Protocol.
- Changes can be accepted
DL-Protocol principles
- Events and marks basics.
- The 'DL-Protocol window' shows the sequence of DL-Protocol events and DL-Protocol marks that have to be set to complete an experiment. Besides, it is possible to set user-defined marks and user-defined events (menu 'Experiment' -> 'Add event' or F4 or STRG-left-click in plot), which will not affect the DL-Protocol and will be stored together with the DL-Protocol marks and events in the .DLD data file. Extra user-defined events are of importance for unpredictable occurrences e.g. when a chamber has to be re-oxygenised, requiring 'open' and 'close' events.
- It is possible to edit/change a user-defined mark or user-defined event into DL-Protocol mark or DL-Protocol event, respectively.
- In most instances, a DL-Protocol event is a titration step of a specific substance according to the DL Protocol. Thereby, the DL-Protocol event demarcates a specific 'state' (in particular a respiratory state, depending on the titrated substance) until the next DL-Protocol event. Within a 'states' a DL-Protocol mark is set to read out the state-specific oxygen flux (or any other signal such as oxygen or fluorescence - has to be defined in the DL-Protocol).
- Events and marks basics.
- Rules. For the correct completion of a DL-Protocol, every single DL-Protocol mark (">> M:") or DL-Protocol event (">> E:") must be set only once in the right order. The exception is a DL-Protocol multi-event ("∗ E:"), which is allowed to be set successively multiple times before proceeding with the next DL-Protocol event in the DL-Protocol sequence. Color-coding of events and marks helps to keep track of the correct process through the DL-Protocol, see 'DL-Protocol colors'.
- Events in action. An HRFR experiment is primarily an event-driven process: DL-Protocol events MUST be set; DL-Protocol marks DON'T NEED to be regarded at first, when running the DL-Protocol. Hence, for practicability, the DL-Protocol marks can be hidden in the list ('DL-Protocol window':
 ) and the user only has to focus on the sequence of the DL-Protocol events. Still, DL-Protocol marks or any user-defined marks can be set at any time during the experimental run.
) and the user only has to focus on the sequence of the DL-Protocol events. Still, DL-Protocol marks or any user-defined marks can be set at any time during the experimental run.
- Events in action. An HRFR experiment is primarily an event-driven process: DL-Protocol events MUST be set; DL-Protocol marks DON'T NEED to be regarded at first, when running the DL-Protocol. Hence, for practicability, the DL-Protocol marks can be hidden in the list ('DL-Protocol window':
- Marks in action For practicability, it is recommended to set the DL-Protocol marks after the experimental procedure has been completed. By definition, a DL-Protocol mark is necessarily linked to a preceding DL-Protocol event, as determined in DL-Protocol sequence. Only when set behind the corresponding DL-Protocol event, The DL-Protocol mark is correctly set and the name is automatically assigned. With DL-Protocol events set in a plot, the DL-Protocol marks can be set in any order, because the automatic mark naming is based on the preceding DL-Protocol event. When setting a second or any further mark in a state (behind a DL-Protocol event), this mark will be recognised as a user-defined mark, automatically named with a single letter (alphabetically, starting with 'a'). The same is true, when a mark is set in the plot behind an DL-Protocol event, where no DL-Protocol mark is defined in the DL-Protocol.
- DL-Protocol colors. In the process of running a DL-Protocol, following the DL Protocol events (>> E: or ∗ E:) and even after when setting DL Protocol marks (>> M:) in the list of the 'DL-Protocol window', events and marks appear in different colors according to the following rules:
Color type indicator Rules, description Yellow events ">> E:" or "∗ E:" Indicates the next DL-Protocol event going to be set according to the DL-Protocol Green event ">> E:" The DL-Protocol event is set/completed in the correct position according to the DL-Protocol. Aqua mulit event "∗ E:" The DL-Protocol multi event is set/completed in the correct position according to the DL-Protocol. As intended, a multi event "∗ E: xxx *"(in auqua) can be consecutively set/clicked multiple times before processing with the next DL-Protocol event (in yellow) in the DL-Protocol.
Light green mark ">> M:" The DL-Protocol mark occurs at the right position, after the associated DL-Protocol event. Red events ">> E:" or "∗ E:" (i) The DL-Protocol event occurs in the wrong position e.g. a later on DL-Protocol event from the DL-Protocol sequence has been set too early. (ii) A scheduled DL-Protocol event is missing in(has been left out) or has been deleted from the processed sequence - can lead to a sequence of red events. (iii) The "">> E:" DL-Protocol event (not a multi-event!) occurs multiple times in the plot. marks ">> M:" (i) The DL-Protocol mark occurs in the wrong place according to the DL-Protocol. (ii) The DL-Protocol mark occurs more than once in the plot. (iii) According tot the rules, a DL-Protocol mark is always necessarily linked to a preceding DL-Protocol event. When this particular DL-Protocol event is coded red, also the associated DL-Protocol mark is considered wrong. White events ">> E:" or "∗ E:" The DL-Protocol event is not set in the plot. marks ">> M:" The DL-Protocol mark is not set in the plot.
Running a DL-Protocol
- Start. Hardware and Software must be setup properly (refer to the O2k-FluoRespirometer manual). DatLab 7 has to be started, connected ('Connect to O2k') and all instrumental- and experimental parameters have to be set.
- Loading a DL-Protocol. A DL-Protocol (either .DLP or DLPU) can be loaded for each chamber in 'Set DL-Protocol / O2 limit'. When the same DL-Protocol is loaded for both chambers, the 'Synchronous DL-Protocol events' mode of operation is available. The loaded DL-protocols for each chamber are displayed in the 'DL-Protocol window' to the right in DatLab 7.
- Running a DL-Protocol. Once the DL-Protocol is loaded, the protocol layout will be displayed on the right side in the 'DL-Protocol window'. The experiment can now be started following the DL Protocol event sequence. For this purpose, left-klick onto the first DL Protocol event in yellow (>> E: xxx) of the DL Protocol and the Event information window will open to set the DL-Protocol event in the plot. When the DL-Protocol event is set correctly according to the protocol sequence it turns into green (>> E: xxx) in the DL-Protocol window list. Proceed with the next yellow DL-Protocol event >> E: yyy at the according time point (after flux has stabilized or experiment specific timing). When a multi-event is encountered the first time to be set it appears in yellow (∗ E: xxx *). After that it appears in auqua (∗ E: xxx *) and can be set multiple times, according to the experimental needs.
Titrations in SUIT protocols
- Stock concentration of substance B in the titration syringe, cB,in [mM] or [µM] as specified.
- Titration volume, v [µl]
- Chamber volume, V [µl]
- Final concentration, cB [mM] or [µM] as specified.
- cB [mM] = cB,in [mM] x v [µl] / V [µl]
- Example: Malate (M), cM,in = 800 mM stock; v = 5 µl titration volume; V = 2,000 µl chamber volume.
- cM [mM] = cM,in [mM] x v [µl] / V [µl] = 800 mM x 5 µl / 2,000 µl = 2 mM