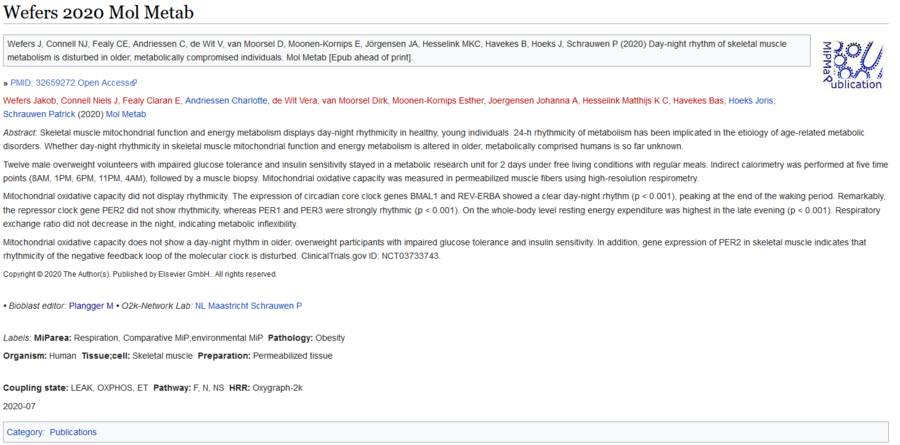
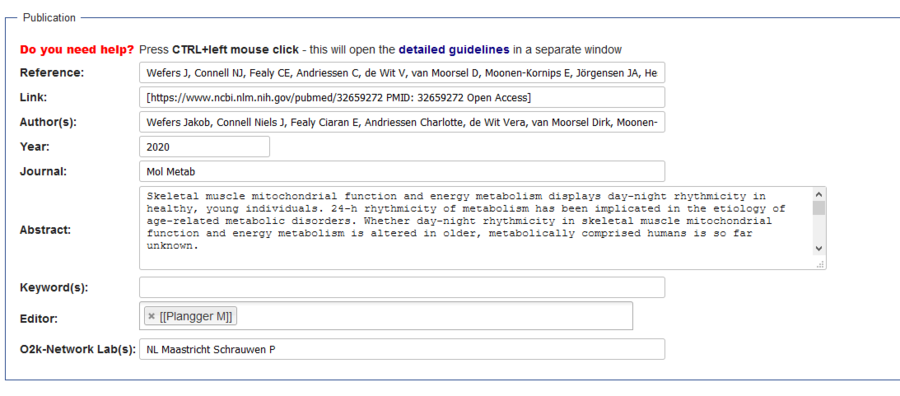
Difference between revisions of "Template talk:Next oroboros ecosystem agenda"
(→Next) |
Timon Alba (talk | contribs) (→Next) |
||
| Line 14: | Line 14: | ||
| width="110" align="center" | [[Image:O2k-Support.jpg|70px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Red-loss of data.png|25px|Warning: loss of data]] | | width="110" align="center" | [[Image:O2k-Support.jpg|70px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Red-loss of data.png|25px|Warning: loss of data]] | ||
|'''Did you know that the proper use of TIP2k-Filter Papers may avoid an unstable O2 slope neg. signal during Instrumental background correction?''' <br> | |'''Did you know that the proper use of TIP2k-Filter Papers may avoid an unstable O2 slope neg. signal during Instrumental background correction?''' <br> | ||
- »[ | - »[[Oxygen_flux_-_instrumental_background#Unstable_O2_slope_neg._signal_in_O2_instrumental_BG|Instrumental background]]« and »[[TIP2k Filter Papers]]« | ||
|} | |} | ||
:::: ''See the scheme'': | :::: ''See the scheme'': | ||
| Line 20: | Line 20: | ||
[[Image:TEMPLATE ALERT.png|none|900px|Agenda/Alerts body text format]] | [[Image:TEMPLATE ALERT.png|none|900px|Agenda/Alerts body text format]] | ||
== MitoPedia alert == | == MitoPedia alert == | ||
::: Example: | |||
:{| | :{| | ||
| width="110" align="center" | [[File:MitoPedia.jpg|left|80px|MitoPedia|link=MitoPedia]] | | width="110" align="center" | [[File:MitoPedia.jpg|left|80px|MitoPedia|link=MitoPedia]] | ||
|''' | |''' Measuring fatty acid oxidation (FAO)-linked respiration requires a low concentration of malate '''<br> | ||
In | In the absence of malate, accumulation of Acetyl-CoA inhibits FAO. <br> | ||
- » | - »'''[[Fatty acid oxidation pathway control state]]'''«, »'''[https://www.bioblast.at/index.php/Gnaiger_2020_BEC_MitoPathways Mitochondrial Pathways: Chapter 5]'''« <br> | ||
- communicated by »[[Cecatto Cristiane]]« and »[[Cardoso Luiza]]« | |||
|} | |} | ||
=== Next === | |||
=== Under construction === | |||
on hold until the page is updated | |||
:{| | :{| | ||
| width=" | | width="100" align="center" |[[File:FAT4BRAIN logo.png|right|80px|link=https://wiki.oroboros.at/index.php/FAT4BRAIN|FAT4BRAIN]] | ||
|''' | |'''A FAO-mito-CNS platform to help fatty acid-related respiration measurements was developed in the FAT4BRAIN project''' <br> | ||
- » [[FAT4BRAIN_FAO-mito-CNS_platform|FAT4BRAIN FAO-mito-CNS platform]] « | |||
- » | |||
- | |||
|} | |} | ||
* | == O2k-Brief alert == | ||
:::* Example: | |||
:{| | :{| | ||
| width=" | | width="100" align="center" | [[File:O2k-brief.png|45px|link=http://wiki.oroboros.at/index.php/O2k-brief|O2k-brief repository]] | ||
|'''Antitumoral properties of triterpenic acid functionalized gold nanoparticles arise from decreasing mitochondrial respiration''' <br> | |||
* Mioc M, Mioc A, Racoviceanu R, Ghiulai R, Prodea A, Milan A, Barbu Tudoran L, Oprean C, Ivan V, Șoica C (2023) The antimelanoma biological assessment of triterpenic acid functionalized gold nanoparticles. [[Mioc 2023 Molecules|»Bioblast link«]] | |||
- | |||
|''' | |||
|} | |} | ||
=== Next === | |||
== O2k-Publications alert == | |||
: | :::* Example: | ||
:{| | :{| | ||
| width="100" align="center" |[[Image:O2k-Publications.jpg|70px|link=http://wiki.oroboros.at/index.php/O2k-Publications:_Topics |O2k-Publications in the MiPMap]] | |||
|'''"Mitochondria–nucleus communication and its integration within the maintenance of cellular homeostasis constitute key determinants of cellular fate and organism health in the context of cellular stress, homeostasis perturbation, and disease"''' <br> | |||
* Temelie M, Talpur R, Dominguez-Prieto M, Dantas Silva A, Cenusa C, Craciun L, Savu DI, Moisoi N (2023) Impaired integrated stress response and mitochondrial integrity modulate genotoxic stress impact and lower the threshold for immune signalling. [[Temelie 2023 Int J Mol Sci|»Bioblast link«]] | |||
:from the O2k-Network »[[UK Leicester Moisoi N]]« | |||
| width="100" align="center" | [[ | |||
|''' | |||
|} | |} | ||
===='''Author names of O2k-Publications/ Brief for the Agenda vs Bioblast''' ==== | ===='''Author names of O2k-Publications/ Brief for the Agenda vs Bioblast''' ==== | ||
__NOTOC__ [[File:Expand.png|right|25px|Click to expand or collaps]] | __NOTOC__ [[File:Expand.png|right|25px|Click to expand or collaps]] | ||
| Line 159: | Line 101: | ||
</div> | </div> | ||
=== Next === | |||
May 22nd | |||
:{| | :{| | ||
| width="100" align="center" |[[Image:O2k-Publications.jpg|70px|link=http://wiki.oroboros.at/index.php/O2k-Publications:_Topics |O2k-Publications in the MiPMap]] | | width="100" align="center" |[[Image:O2k-Publications.jpg|70px|link=http://wiki.oroboros.at/index.php/O2k-Publications:_Topics |O2k-Publications in the MiPMap]] | ||
|''' | |'''"We have identified a circadian clock phase-dependent change in Complex I coupling efficiency, demonstrating for the first time a mitochondria–clock interaction in the hooded seal."''' <br> | ||
* Ciccone C, Kante F, Folkow LP, Hazlerigg DG, West AC, Wood SH (2024) Circadian coupling of mitochondria in a deep-diving mammal. [[Ciccone 2024 J Exp Biol|»Bioblast link«]] | |||
* | :from the O2k-Network »[[NO Tromsoe Wood SH]]« | ||
|} | |} | ||
May 9th | |||
:{| | :{| | ||
| width="100" align="center" |[[Image:O2k-Publications.jpg|70px|link=http://wiki.oroboros.at/index.php/O2k-Publications:_Topics |O2k-Publications in the MiPMap]] | | width="100" align="center" |[[Image:O2k-Publications.jpg|70px|link=http://wiki.oroboros.at/index.php/O2k-Publications:_Topics |O2k-Publications in the MiPMap]] | ||
|''' | |'''"The methodology exploited in this work opens the possibility to extrapolate the photosynthetic functionality of microalgae in dense cultures typical of industrial cultivation plants already at the lab-scale"''' <br> | ||
:from the O2k-Network »[[ | * Vera-Vives AM, Michelberger T, Morosinotto T, Perin G (2024) Assessment of photosynthetic activity in dense microalgae cultures using oxygen production. Plant Physiol Biochem [[Vera-Vives 2024 Plant Physiol Biochem|»Bioblast link«]] | ||
:from the O2k-Network »[[IT Padova Morosinotto T]]« | |||
|} | |} | ||
== MiPNet == | |||
:::* Example: | |||
:{| | |||
| width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Blue-info.png|25px|Info: improve]] | |||
|''' "No bubbles should be trapped in the electrolyte reservoir after membrane application and mounting"'''<br> | |||
- »'''[[MiPNet28.10 SmartPOS-service]]'''«<br> | |||
- communicated by »[[Timon-Gomez Alba]]« and »[[]]« | |||
|} | |} | ||
== | === Next === | ||
May 2nd | |||
:{| | :{| | ||
| width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Blue-info.png|25px|Info: improve]] | | width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Blue-info.png|25px|Info: improve]] | ||
| ''' | |''' "In respirometric studies on blood cells, the purity of cell preparation should always be emphasized and documented." '''<br> | ||
- »'''[[MiPNet21.17 BloodCellsIsolation]]'''«<br> | |||
- » [ | - communicated by »[[Timon-Gomez Alba]]« and »[[Baglivo Eleonora]]« | ||
|} | |} | ||
=== Next === | |||
May 16th | |||
:{| | :{| | ||
| width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Blue-info.png|25px|Info: improve]] | | width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Blue-info.png|25px|Info: improve]] | ||
| ''' | |''' "The sample holder protects (...) fragile samples from potential damage caused by stirring the medium."'''<br> | ||
- »'''[[MiPNet27.07 Sample Holder]]'''«<br> | |||
- »[[ | - communicated by »[[Willis Jaime R]]« and »[[Timon-Gomez Alba]]« | ||
|} | |||
== O2k-Open Support alert == | |||
- » [[ | :::*Examples: | ||
:{| | |||
| width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Yellow-take care.png|25px|Info: take care]] | |||
|'''Does your polarographic oxygen sensor (POS) need a deep clean?'''<br> | |||
A POS with a very dark anode might need several rounds of ammonia cleaning, possibly including prolonged exposure overnight to restore performance specifications. | |||
<br> | |||
- »'''[[MiPNet19.18B POS-service|POS service]]'''«<br> | |||
- communicated by »[[Cecatto Cristiane]]« and »[[Leo Elettra]]« | |||
|} | |||
:{| | |||
| width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Red-loss of data.png|25px|Warning: loss of data]] | |||
|'''Did you know that a humid environment and/or liquid spillage may lead to a high raw POS signal [V]?'''<br> | |||
See how to clean and dry the electrical connections of the OroboPOS-Connector.<br> | |||
- »'''[[OroboPOS-Connector#Cleaning_the_electrical_connections|Cleaning the electrical connections]]'''«<br> | |||
- communicated by »[[Cecatto Cristiane]]« and »[[Baglivo Eleonora]]« | |||
|} | |} | ||
:{| | :{| | ||
| width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Blue-info.png|25px|Info: improve]] | | width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Blue-info.png|25px|Info: improve]] | ||
| ''' | |''' Unusual stirrer bar rotation? '''<br> | ||
A “jumping” stirrer or a slow rotation of the stirrer bar may indicate a problem either with the stirrer itself or with the cleaning of the glass chamber. | |||
- »[ | <br> | ||
- »'''[https://wiki.oroboros.at/index.php/Stirrer-Bar%5Cwhite_PVDF%5C15x6_mm#Technical_support Stirrer bar - technical support]'''«<br> | |||
- communicated by »[[Baglivo Eleonora]]« and »[[Grings Mateus]]« | |||
|} | |} | ||
=== Next === | |||
May 27th | |||
:{| | :{| | ||
| width="110" align="center" | [[ | | width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Yellow-take care.png|25px|Info: take care]] | ||
|''' | |'''Same experimental conditions, but different O<sub>2</sub> fluxes in the left and right chambers?'''<br> | ||
Check for instrumental and operational factors that could be causing O<sub>2</sub> flux differences between the O2k-chambers. | |||
- »'''[[ | <br> | ||
- »'''[[Different O2 fluxes in left and right chamber]]'''«<br> | |||
- communicated by »[[Timon-Gomez Alba]]« and »[[Baglivo Eleonora]]« | |||
|} | |} | ||
May 13th | |||
:{| | :{| | ||
| width="110" align="center" | [[ | | width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Yellow-take care.png|25px|Info: take care]] | ||
|''' | |'''Do you see fluctuations of O<sub>2</sub> slope negative, even in the absence of sample, following a pattern?'''<br> | ||
Placing the O2k under direct airflow of an air conditioning equipment may lead to disturbances in the O<sub>2</sub> slope negative. | |||
- » | <br> | ||
- »'''[[Oxygen_flux_-_instrumental_background#Fluctuations_of_O2_slope_negative|Fluctuations of O<sub>2</sub> slope negative]]'''«<br> | |||
- communicated by »[[Cardoso Luiza]]« and »[[Grings Mateus]]« | |||
|} | |} | ||
or | |||
:{| | :{| | ||
| width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File: | | width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Yellow-take care.png|25px|Info: take care]] | ||
| ''' | |'''Do you see fluctuations of O<sub>2</sub> slope negative following a pattern similar to the Peltier power, but without block temperature variation?'''<br> | ||
Placing the O2k under direct airflow of an air conditioning equipment may lead to disturbances in the O<sub>2</sub> slope negative. | |||
- » [[ | <br> | ||
- »'''[[Oxygen_flux_-_instrumental_background#Fluctuations_of_O2_slope_negative|Fluctuations of O<sub>2</sub> slope negative]]'''«<br> | |||
- communicated by »[[Cardoso Luiza]]« and »[[Grings Mateus]]« | |||
|} | |} | ||
== O2k-Feedback alert == | |||
:::* Example (if the person has a profile, link the name to the profile with [[Cecatto C| Cristiane Cecatto]]: | |||
:{| | |||
| width="110" align="center" | [[Image:O2k-Feedback.jpg|60px|link=https://wiki.oroboros.at/index.php/O2k-Feedback |O2k-Feedback]] | |||
|''' ''“Thank you very much for your thorough help. I appreciate the time you took to provide me with all the answers and supportive literature to realize the potential of the O2k-respirometer in bioenergetics research”'''''<br> | |||
Badr Ibrahim, PhD from Betterhumans Inc., US | |||
<br> | |||
- »'''[[O2k-Feedback]]'''«<br> | |||
|} | |||
:{| | :{| | ||
| width="110" align="center" | [[Image:O2k- | | width="110" align="center" | [[Image:O2k-Feedback.jpg|60px|link=https://wiki.oroboros.at/index.php/O2k-Feedback |O2k-Feedback]] | ||
|''' | |''' ''“Thank you very much for your thorough help. I appreciate the time you took to provide me with all the answers and supportive literature to realize the potential of the O2k-respirometer in bioenergetics research”'''''<br> | ||
Badr Ibrahim, PhD from the O2k-Network »[[UK Leicester Moisoi N]]«<br> | |||
- »[[ | - »'''[[O2k-Feedback]]'''«<br> | ||
|} | |} | ||
* | === Next === | ||
== MitoFit Preprints == | |||
:::* Example: | |||
:{| | :{| | ||
| width=" | | width="100" align="center"|[[Image:BEC-logo.png|100px|link=https://www.bioenergetics-communications.org/index.php/bec |Bioenergetics Communications]] | ||
|''' | |'''From preprint to BEC manuscript with published Open Peer Reviews:'''<br> | ||
*Komlódi T, Sobotka O, Gnaiger E (2021) Facts and artefacts on the oxygen dependence of hydrogen peroxide production using Amplex UltraRed. Bioenerg Commun 2021.4. [[doi:10.26124/BEC:2021-0004]] | |||
:from the O2k-Network »[[AT Innsbruck Oroboros]]« | |||
|} | |} | ||
== | === Next === | ||
== Under discussion == | |||
This one needs an update of the Bioblast page and clarification of MiPNet06.03 | |||
:{| | :{| | ||
| width=" | | width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Blue-info.png|25px|Info: improve]] | ||
| | |''' Did you enter the correct oxygen solubility factor in DatLab?'''<br> | ||
The oxygen solubility factor of the incubation medium, F<sub>M</sub>, expresses the effect of the salt concentration on oxygen solubility relative to pure water. The correct oxygen solubility factor of your experimental medium or buffer needs to be entered in DatLab for proper calculation of the oxygen concentration. | |||
<br> | |||
- »'''[[Oxygen solubility factor|Oxygen solubility factor]]'''«<br> | |||
- communicated by »[[Schmitt Sabine]]« and »[[ ]]« | |||
|} | |} | ||
This one needs a BioBlast link: | |||
:: | :{| | ||
| width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Blue-info.png|25px|Info: improve]] | |||
|''' When filling in the volume of uncoupler in the DatLab mark or the excel template, should one enter the total volume added or the volume to get to the highest flux?'''<br> | |||
The volume added until the mark, equal to the highest flux, should be entered. Anything titrated after this mark should be summed to the other volumes before the next mark because this will be considered to calculate the sample dilution. | |||
<br> | |||
- »'''[[LINK|LINK]]''' and '''[[MiPNet24.06_Oxygen_flux_analysis_-_DatLab_7.4|O<sub>2</sub> flux analysis with DatLab 7.4]]'''«<br> | |||
- communicated by »[[Cardoso Luiza]]« and »[[ ]]« | |||
|} | |||
::: | == Removed for correction == | ||
* '''Nov-30 Mo''' | |||
:{| | |||
| width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Blue-info.png|25px|Info: improve]] | |||
|'''Noisy oxygen signal? Check the OroboPOS and OroboPOS-Connector to clean electrical connections.'''<br> | |||
Contaminating grease, salt crystals and moisture can be removed with a dry paper cloth or a paper cloth moistened with water and then absolute ethanol. <br> | |||
- »[[OroboPOS-Connector#Cleaning_the_electrical_connections |Cleaning the electrical connections]]« <br> | |||
- communicated by »[[Schmitt Sabine]]« and »[[Komlodi Timea]]« | |||
|} | |||
WGT recommended not to use water for cleaning. Erich introduced water for cleaning to remove salt crystals. | |||
: | :{| | ||
| width="110" align="center" | [[Image:O2k-Support.jpg|60px|link=http://wiki.oroboros.at/index.php/O2k-Open_Support |O2k-Open Support]][[File:Red-loss of data.png|25px|Warning: loss of data]] | |||
|'''Do you perform the ''leaky chamber test'' after chamber assembly? '''<br> | |||
An unnoticed leak from the chamber may affect your data, therefore we recommend performing the ''Leaky chamber test'' - especially after chamber reassembly. <br> | |||
- »[[Talk:O2k-Chamber_MitoPedia#Leaky_Chamber|Leaky chamber test]]« and » https://intern.oroboros.at/index.php/Overnight_test_after_O2k-chamber_assembly« | |||
|} | |||
Latest revision as of 15:19, 2 May 2024
- Antunes Diana (talk) 08:58, 5 May 2020 (CEST) Format for new Agenda/Alerts entries:
{|
| logos
| title in bold <br>
- link
|}
e.g.,


Did you know that the proper use of TIP2k-Filter Papers may avoid an unstable O2 slope neg. signal during Instrumental background correction?
- »Instrumental background« and »TIP2k Filter Papers«
- See the scheme:
MitoPedia alert
- Example:
Measuring fatty acid oxidation (FAO)-linked respiration requires a low concentration of malate
In the absence of malate, accumulation of Acetyl-CoA inhibits FAO.
- »Fatty acid oxidation pathway control state«, »Mitochondrial Pathways: Chapter 5«
- communicated by »Cecatto Cristiane« and »Cardoso Luiza«
Next
Under construction
on hold until the page is updated
A FAO-mito-CNS platform to help fatty acid-related respiration measurements was developed in the FAT4BRAIN project
O2k-Brief alert
- Example:

Antitumoral properties of triterpenic acid functionalized gold nanoparticles arise from decreasing mitochondrial respiration
- Mioc M, Mioc A, Racoviceanu R, Ghiulai R, Prodea A, Milan A, Barbu Tudoran L, Oprean C, Ivan V, Șoica C (2023) The antimelanoma biological assessment of triterpenic acid functionalized gold nanoparticles. »Bioblast link«
Next
O2k-Publications alert
- Example:

"Mitochondria–nucleus communication and its integration within the maintenance of cellular homeostasis constitute key determinants of cellular fate and organism health in the context of cellular stress, homeostasis perturbation, and disease"
- Temelie M, Talpur R, Dominguez-Prieto M, Dantas Silva A, Cenusa C, Craciun L, Savu DI, Moisoi N (2023) Impaired integrated stress response and mitochondrial integrity modulate genotoxic stress impact and lower the threshold for immune signalling. »Bioblast link«
- from the O2k-Network »UK Leicester Moisoi N«
Author names of O2k-Publications/ Brief for the Agenda vs Bioblast
Guidelines to update a reference
- When creating an O2k-Publication/Brief alert, it is the responsibility of the author to check if the reference is accordingly to the correct guidelines:
- Correct format for a person called “First Name Surname”:
- In the gray box should be “Surname FN” - From where references to be used as O2k-Publication/Brief alerts are copied from.
- In the links should be “ Surname First N”
- How to edit:
- A) Check if all authors with profiles in Bioblast (blue links) have their profiles with full name. If they have, proceed to next step (B). If not, you need to move it following the steps “How to move a profile” below.
- B) Click in “Edit with form”.
- C) The gray box is edited in “Reference” and the links are changed in “Author(s)”. Change it accordingly and click in “save page” at the end of the page.
- How to edit:
How to move a profile (instructions sent by Marija):
So let’s take for example my page: Beno M [1]
- As you can see there is a redirect from Beno Marija to Beno M. What has to be done is to switch the content of those pages. Here are the steps:
- 1. Go to e.g. “Beno M” on edit
- 2. Click on “Move”
- 3. Change the new title to the full name of the person. In my case it will be Beno M to Beno Marija
- As you can see there is a redirect from Beno Marija to Beno M. What has to be done is to switch the content of those pages. Here are the steps:
- 4. Now it should look like that:
- 5. Click on “Move” and leave all the ticked boxes
- 6.Done
- After that you can continue in step B.
- If you have any doubt you can contact Cristiane, Marija or Mario.
Next
May 22nd

"We have identified a circadian clock phase-dependent change in Complex I coupling efficiency, demonstrating for the first time a mitochondria–clock interaction in the hooded seal."
- Ciccone C, Kante F, Folkow LP, Hazlerigg DG, West AC, Wood SH (2024) Circadian coupling of mitochondria in a deep-diving mammal. »Bioblast link«
- from the O2k-Network »NO Tromsoe Wood SH«
May 9th

"The methodology exploited in this work opens the possibility to extrapolate the photosynthetic functionality of microalgae in dense cultures typical of industrial cultivation plants already at the lab-scale"
- Vera-Vives AM, Michelberger T, Morosinotto T, Perin G (2024) Assessment of photosynthetic activity in dense microalgae cultures using oxygen production. Plant Physiol Biochem »Bioblast link«
- from the O2k-Network »IT Padova Morosinotto T«
MiPNet
- Example:


"No bubbles should be trapped in the electrolyte reservoir after membrane application and mounting"
- »MiPNet28.10 SmartPOS-service«
- communicated by »Timon-Gomez Alba« and »[[]]«
Next
May 2nd


"In respirometric studies on blood cells, the purity of cell preparation should always be emphasized and documented."
- »MiPNet21.17 BloodCellsIsolation«
- communicated by »Timon-Gomez Alba« and »Baglivo Eleonora«
Next
May 16th


"The sample holder protects (...) fragile samples from potential damage caused by stirring the medium."
- »MiPNet27.07 Sample Holder«
- communicated by »Willis Jaime R« and »Timon-Gomez Alba«
O2k-Open Support alert
- Examples:


Does your polarographic oxygen sensor (POS) need a deep clean?
A POS with a very dark anode might need several rounds of ammonia cleaning, possibly including prolonged exposure overnight to restore performance specifications.
- »POS service«
- communicated by »Cecatto Cristiane« and »Leo Elettra«


Did you know that a humid environment and/or liquid spillage may lead to a high raw POS signal [V]?
See how to clean and dry the electrical connections of the OroboPOS-Connector.
- »Cleaning the electrical connections«
- communicated by »Cecatto Cristiane« and »Baglivo Eleonora«


Unusual stirrer bar rotation?
A “jumping” stirrer or a slow rotation of the stirrer bar may indicate a problem either with the stirrer itself or with the cleaning of the glass chamber.
- »Stirrer bar - technical support«
- communicated by »Baglivo Eleonora« and »Grings Mateus«
Next
May 27th


Same experimental conditions, but different O2 fluxes in the left and right chambers?
Check for instrumental and operational factors that could be causing O2 flux differences between the O2k-chambers.
- »Different O2 fluxes in left and right chamber«
- communicated by »Timon-Gomez Alba« and »Baglivo Eleonora«
May 13th


Do you see fluctuations of O2 slope negative, even in the absence of sample, following a pattern?
Placing the O2k under direct airflow of an air conditioning equipment may lead to disturbances in the O2 slope negative.
- »Fluctuations of O2 slope negative«
- communicated by »Cardoso Luiza« and »Grings Mateus«
or


Do you see fluctuations of O2 slope negative following a pattern similar to the Peltier power, but without block temperature variation?
Placing the O2k under direct airflow of an air conditioning equipment may lead to disturbances in the O2 slope negative.
- »Fluctuations of O2 slope negative«
- communicated by »Cardoso Luiza« and »Grings Mateus«
O2k-Feedback alert
- Example (if the person has a profile, link the name to the profile with Cristiane Cecatto:

“Thank you very much for your thorough help. I appreciate the time you took to provide me with all the answers and supportive literature to realize the potential of the O2k-respirometer in bioenergetics research”
Badr Ibrahim, PhD from Betterhumans Inc., US
- »O2k-Feedback«

“Thank you very much for your thorough help. I appreciate the time you took to provide me with all the answers and supportive literature to realize the potential of the O2k-respirometer in bioenergetics research”
Badr Ibrahim, PhD from the O2k-Network »UK Leicester Moisoi N«
- »O2k-Feedback«
Next
MitoFit Preprints
- Example:

From preprint to BEC manuscript with published Open Peer Reviews:
- Komlódi T, Sobotka O, Gnaiger E (2021) Facts and artefacts on the oxygen dependence of hydrogen peroxide production using Amplex UltraRed. Bioenerg Commun 2021.4. doi:10.26124/BEC:2021-0004
- from the O2k-Network »AT Innsbruck Oroboros«
Next
Under discussion
This one needs an update of the Bioblast page and clarification of MiPNet06.03


Did you enter the correct oxygen solubility factor in DatLab?
The oxygen solubility factor of the incubation medium, FM, expresses the effect of the salt concentration on oxygen solubility relative to pure water. The correct oxygen solubility factor of your experimental medium or buffer needs to be entered in DatLab for proper calculation of the oxygen concentration.
- »Oxygen solubility factor«
- communicated by »Schmitt Sabine« and »[[ ]]«
This one needs a BioBlast link:


When filling in the volume of uncoupler in the DatLab mark or the excel template, should one enter the total volume added or the volume to get to the highest flux?
The volume added until the mark, equal to the highest flux, should be entered. Anything titrated after this mark should be summed to the other volumes before the next mark because this will be considered to calculate the sample dilution.
- »LINK and O2 flux analysis with DatLab 7.4«
- communicated by »Cardoso Luiza« and »[[ ]]«
Removed for correction
- Nov-30 Mo


Noisy oxygen signal? Check the OroboPOS and OroboPOS-Connector to clean electrical connections.
Contaminating grease, salt crystals and moisture can be removed with a dry paper cloth or a paper cloth moistened with water and then absolute ethanol.
- »Cleaning the electrical connections«
- communicated by »Schmitt Sabine« and »Komlodi Timea«
WGT recommended not to use water for cleaning. Erich introduced water for cleaning to remove salt crystals.


Do you perform the leaky chamber test after chamber assembly?
An unnoticed leak from the chamber may affect your data, therefore we recommend performing the Leaky chamber test - especially after chamber reassembly.
- »Leaky chamber test« and » https://intern.oroboros.at/index.php/Overnight_test_after_O2k-chamber_assembly«