Gnaiger 2023 MitoFit CII: Difference between revisions
No edit summary |
No edit summary |
||
| (354 intermediate revisions by 3 users not shown) | |||
| Line 1: | Line 1: | ||
{{Publication | {{Publication | ||
|title=Gnaiger E (2023) Complex II ambiguities ― FADH<sub>2</sub> in the electron transfer system. MitoFit Preprints 2023.3. https://doi.org/10.26124/mitofit:2023-0003 | |title=Gnaiger E (2023) Complex II ambiguities ― FADH<sub>2</sub> in the electron transfer system. MitoFit Preprints 2023.3.v6. https://doi.org/10.26124/mitofit:2023-0003.v6 - ''' [[Gnaiger 2024 J Biol Chem |''Published 2023-11-22 J Biol Chem (2024)'']] | ||
|info=MitoFit Preprints 2023.3. [[File:MitoFit Preprints pdf.png|left|160px|link=https://wiki.oroboros.at/images/a/ae/Gnaiger_2023_MitoFit_CII.pdf|MitoFit pdf]] [https://wiki.oroboros.at/images/a/ae/Gnaiger_2023_MitoFit_CII.pdf Complex II ambiguities ― FADH<sub>2</sub> in the electron transfer system]<br/> | |info=MitoFit Preprints 2023.3.v6. [[File:MitoFit Preprints pdf.png|left|160px|link=https://wiki.oroboros.at/images/a/ae/Gnaiger_2023_MitoFit_CII.pdf|MitoFit pdf]] [https://wiki.oroboros.at/images/a/ae/Gnaiger_2023_MitoFit_CII.pdf Complex II ambiguities ― FADH<sub>2</sub> in the electron transfer system]<br/> | ||
|authors=Gnaiger Erich | |authors=Gnaiger Erich | ||
|year=2023 | |year=2023 | ||
|journal=MitoFit Prep | |journal=MitoFit Prep | ||
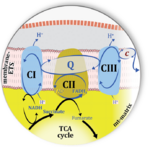
|abstract=[[File:CII-ambiguities Graphical abstract.png| | |abstract=[[File:CII-ambiguities Graphical abstract.png|150px|left]] | ||
The | ::: Gnaiger E (2024) Complex II ambiguities ― FADH<sub>2</sub> in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 | ||
|keywords=coenzyme Q junction; Complex CII; electron transfer system; fatty acid oxidation; flavin adenine dinucleotide; | ::: <small>Version 6 (v6) 2023-06-21 </small> | ||
succinate dehydrogenase; tricarboxylic acid cycle | ::: <small>Version 5 (v5) 2023-05-31, (v4) 2023-05-12, (v3) 2023-05-04, (v2) 2023-04-04, (v1) 2023-03-24 - [https://wiki.oroboros.at/index.php/File:Gnaiger_2023_MitoFit_CII.pdf »Link to all versions«]</small> | ||
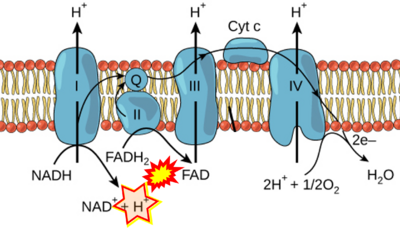
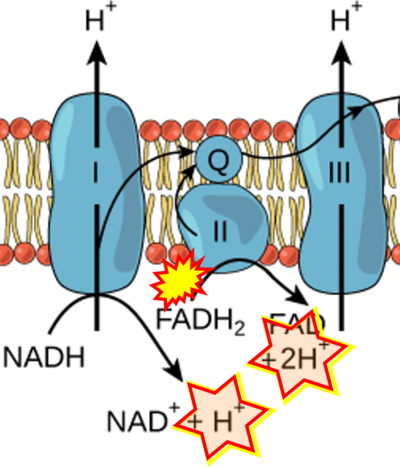
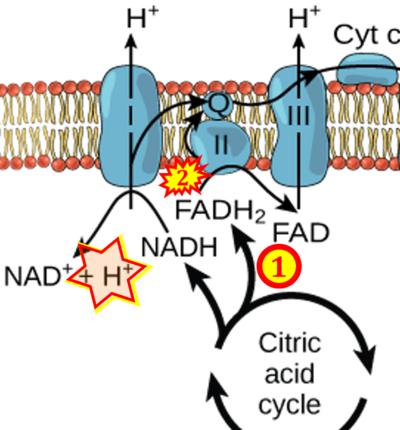
The prevailing notion that reduced cofactors NADH and FADH<sub>2</sub> transfer electrons from the tricarboxylic acid cycle to the mitochondrial electron transfer system creates ambiguities regarding respiratory Complex II (CII). The succinate dehydrogenase subunit SDHA of CII oxidizes succinate and reduces the covalently bound prosthetic group FAD to FADH<sub>2</sub> in the canonical forward tricarboxylic acid cycle. However, several graphical representations of the electron transfer system depict FADH<sub>2</sub> in the mitochondrial matrix as a substrate to be oxidized by CII. This leads to the false conclusion that FADH<sub>2</sub> from the β-oxidation cycle in fatty acid oxidation feeds electrons into CII. In reality, dehydrogenases of fatty acid oxidation channel electrons to the coenzyme Q-junction but not through CII. The ambiguities surrounding Complex II in the literature and educational resources call for quality control, to secure scientific standards in current communications of bioenergetics, and ultimately support adequate clinical applications. This review aims to raise awareness of the inherent '''[[ambiguity crisis]]''', complementing efforts to address the well-acknowledged issues of credibility and reproducibility. | |||
<br> | |||
|keywords=[[coenzyme]]; [[cofactor]]; [[prosthetic group]]; coenzyme Q junction, Q-junction; Complex II, CII; [[H+-linked electron transfer |H<sup>+</sup>-linked electron transfer]]; [[electron transfer system]], ETS; [[matrix-ETS]]; [[membrane-ETS]]; fatty acid oxidation, FAO; flavin adenine dinucleotide, FAD/FADH<sub>2</sub>; nicotinamide adenine dinucleotide, NAD<sup>+</sup>/NADH; succinate dehydrogenase, SDH; tricarboxylic acid cycle, TCA; [[substrate]]; [[Gibbs force]] | |||
|mipnetlab=AT Innsbruck Oroboros | |mipnetlab=AT Innsbruck Oroboros | ||
}} | }} | ||
__TOC__ | |||
::::'''» ''Links:''''' [[Ambiguity crisis]], [[Complex II ambiguities]], [[:Category:Ambiguity crisis - NAD and H+ |Complex I and hydrogen ion ambiguities in the electron transfer system]] | |||
:::: '''Acknowledgements''': I thank [[Cardoso Luiza HD |Luiza H.D. Cardoso]], [[Schmitt Sabine |Sabine Schmitt]], and [[Donnelly Chris |Chris Donnelly]] for stimulating discussions, and [[Cocco Paolo |Paolo Cocco]] for expert help on the graphical abstract and Figures 1d and e. The constructive comments of an anonymous reviewer (J Biol Chem) are explicitly acknowledged. Contribution to the European Union’s Horizon 2020 research and innovation program Grant 857394 ([[FAT4BRAIN]]). | |||
== Additions to 312 references on CII-ambiguities after publication of JBC 2024 == | |||
Last update 2023-12-19 | |||
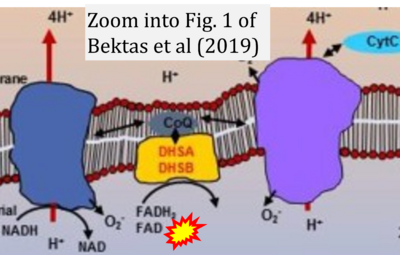
:::::: [[File:Bektas 2019 Aging (Albany NY) CORRECTION.png|400px|link=Bektas 2019 Aging (Albany NY)]] | |||
:::: '''#1''' Bektas A, Schurman SH, Gonzalez-Freire M, Dunn CA, Singh AK, Macian F, Cuervo AM, Sen R, Ferrucci L (2019) Age-associated changes in human CD4+ T cells point to mitochondrial dysfunction consequent to impaired autophagy. '''Aging (Albany NY)''' 11:9234-63. - [[Bektas 2019 Aging (Albany NY) |»Bioblast link«]] | |||
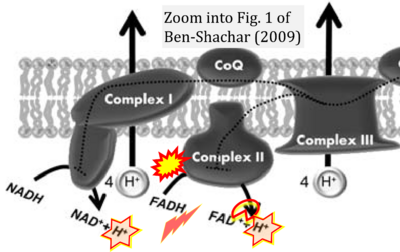
:::::: [[File:Ben-Shachar 2009 J Neural Transm (Vienna) CORRECTION.png|400px|link=Ben-Shachar 2009 J Neural Transm (Vienna)]] | |||
:::: '''#2''' Ben-Shachar D (2009) The interplay between mitochondrial complex I, dopamine and Sp1 in schizophrenia. '''J Neural Transm (Vienna)''' 116:1383-96. - [[Ben-Shachar 2009 J Neural Transm (Vienna) |»Bioblast link«]] | |||
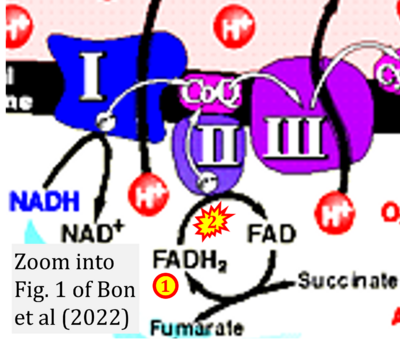
:::::: [[File:Bon 2022 J Clin Case Rep Stud CORRECTION.png|400px|link=Bon 2022 J Clin Case Rep Stud]] | |||
:::: '''#3''' Bon E, Maksimovich NY, Dremza IK (2022) Alendronate-induced nephropathy. '''J Clin Case Rep Stud''' 3. - [[Bon 2022 J Clin Case Rep Stud |»Bioblast link«]] | |||
:::::: [[File:Elsaeed 2021 Medicine Updates CORRECTION.png|400px|link=Elsaeed 2021 Medicine Updates]] | |||
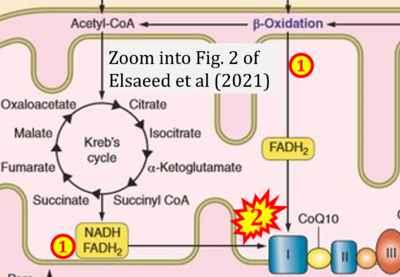
:::: '''#4''' Elsaeed EM, Hamad A, Erfan OS, Elshahat M, Ebrahim F (2021) Role played by hippocampal apoptosis, autophagy and necroptosis in pathogenesis of diabetic cognitive dysfunction: a review of literature. '''Medicine Updates''' 6:41-63. - [[Elsaeed 2021 Medicine Updates |»Bioblast link«]] | |||
:::: | :::::: [[File:Facucho-Oliveira 2009 Stem Cell Rev Rep CORRECTION.png|400px|link=Facucho-Oliveira 2009 Stem Cell Rev Rep]] | ||
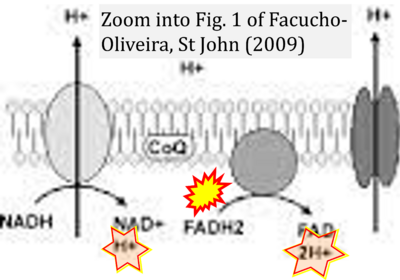
:::: '''#5''' Facucho-Oliveira JM, St John JC (2009) The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. '''Stem Cell Rev Rep''' 5:140-58. - [[Facucho-Oliveira 2009 Stem Cell Rev Rep |»Bioblast link«]] | |||
:::::: [[File:Iqbal 2014 Springer, New York CORRECTION.png|400px|link=Iqbal 2014 Springer, New York]] | |||
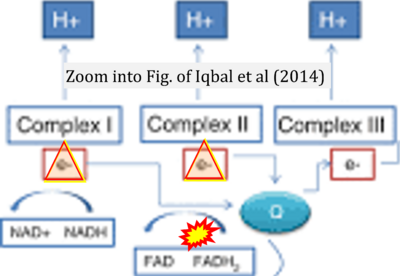
:::: '''#6''' Iqbal T, Welsby PJ, Howarth FC, Bidasee K, Adeghate E, Singh J (2014) Effects of diabetes-induced hyperglycemia in the heart: biochemical and structural slterations. In: Turan B, Dhalla N (eds) Diabetic cardiomyopathy. Advances in biochemistry in health and disease 9. '''Springer''', New York. - [[Iqbal 2014 Springer, New York |»Bioblast link«]] | |||
:::: | :::::: [[File:Keogh 2015 Biochim Biophys Acta CORRECTION.png|400px|link=Keogh 2015 Biochim Biophys Acta]] | ||
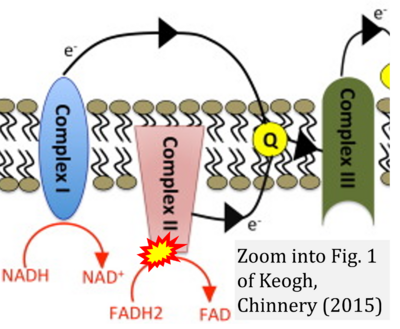
:::: '''#7''' Keogh MJ, Chinnery PF (2015) Mitochondrial DNA mutations in neurodegeneration. '''Biochim Biophys Acta''' 1847:1401-11. - [[Keogh 2015 Biochim Biophys Acta |»Bioblast link«]] | |||
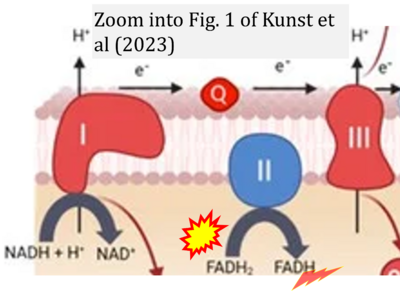
:::: | :::::: [[File:Kunst 2023 Biomedicines CORRECTION.png|400px|link=Kunst 2023 Biomedicines]] | ||
:::::: | :::: '''#8''' Kunst C, Schmid S, Michalski M, Tümen D, Buttenschön J, Müller M, Gülow K (2023) The influence of gut microbiota on oxidative stress and the immune system. '''Biomedicines''' 11:1388. - [[Kunst 2023 Biomedicines |»Bioblast link«]] | ||
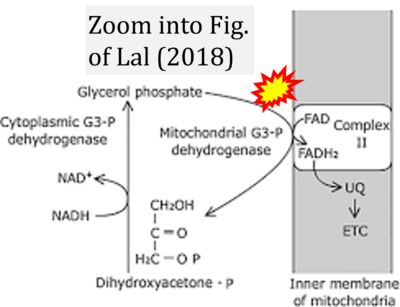
= | :::::: [[File:Lal 2018 Springer CORRECTION.png|400px|link=Lal 2018 Springer]] | ||
:::: '''#9''' Lal MA (2018) Respiration. In: Bhatla SC, Lal MA (eds) Plant physiology, development and metabolism. '''Springer''', Singapore:253-314. - [[Lal 2018 Springer |»Bioblast link«]] | |||
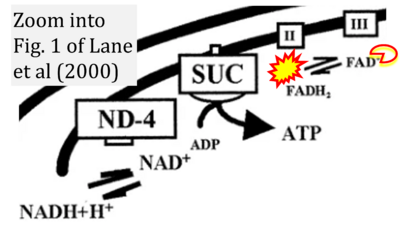
:: | :::::: [[File:Lane 2000 Pediatr Res CORRECTION.png|400px|link=Lane 2000 Pediatr Res]] | ||
:::: | :::: '''#10''' Lane RH, Tsirka AE, Gruetzmacher EM (2000) Uteroplacental insufficiency alters cerebral mitochondrial gene expression and DNA in fetal and juvenile rats. '''Pediatr Res''' 47:792-7. - [[Lane 2000 Pediatr Res |»Bioblast link«]] | ||
::::''' | |||
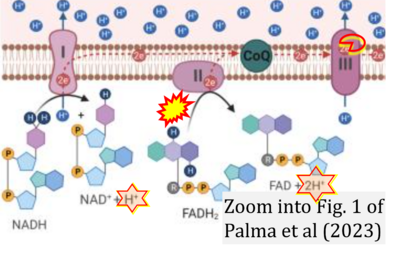
:::::: [[File: | :::::: [[File:Palma 2023 Oncogene CORRECTION.png|400px|link=Palma 2023 Oncogene]] | ||
:::: | :::: '''#11''' Palma FR, Gantner BN, Sakiyama MJ, Kayzuka C, Shukla S, Lacchini R, Cunniff B, Bonini MG (2023) ROS production by mitochondria: function or dysfunction? '''Oncogene'''. - [[Palma 2023 Oncogene |»Bioblast link«]] | ||
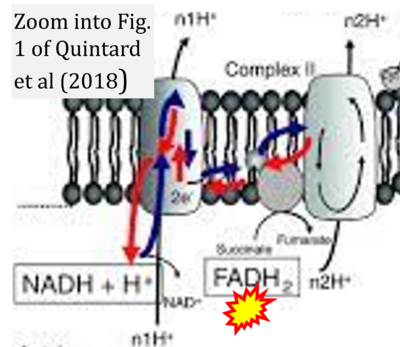
:::::: [[File: | :::::: [[File:Quintard 2018 Springer, Cham CORRECTION.png|400px|link=Quintard 2018 Springer, Cham]] | ||
:::: | :::: '''#12''' Quintard H, Fontaine E, Ichai C (2018) Energy metabolism: from the organ to the cell. In: Ichai, C., Quintard, H., Orban, JC. (eds) Metabolic Disorders and Critically Ill Patients. '''Springer''', Cham. - [[Quintard 2018 Springer, Cham |»Bioblast link«]] | ||
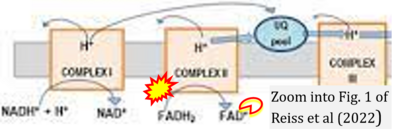
:::: [[File: | :::::: [[File:Reiss 2022 Exp Gerontol CORRECTION.png|400px|link=Reiss 2022 Exp Gerontol]] | ||
:::: | :::: '''#13''' Reiss AB, Ahmed S, Dayaramani C, Glass AD, Gomolin IH, Pinkhasov A, Stecker MM, Wisniewski T, De Leon J (2022) The role of mitochondrial dysfunction in Alzheimer's disease: A potential pathway to treatment. '''Exp Gerontol''' 164:111828. - [[Reiss 2022 Exp Gerontol |»Bioblast link«]] | ||
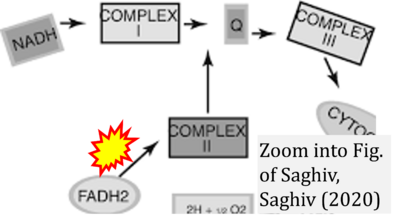
:::::: [[File: | :::::: [[File:Saghiv 2020 Springer, Cham CORRECTION.png|400px|link=Saghiv 2020 Springer, Cham]] | ||
:::: | :::: '''#14''' Saghiv MS, Sagiv MS (2020) Metabolism. In: Basic Exercise Physiology. '''Springer''', Cham. - [[Saghiv 2020 Springer, Cham |»Bioblast link«]] | ||
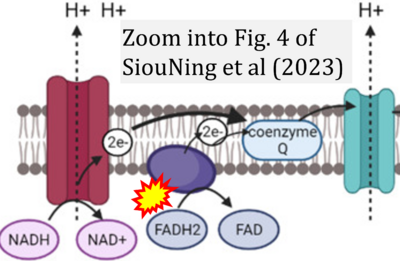
:::::: [[File: | :::::: [[File:SiouNing 2023 Molecules CORRECTION.png|400px|link=SiouNing 2023 Molecules]] | ||
:::: | :::: '''#15''' SiouNing AS, Seong TS, Kondo H, Bhassu S (2023) MicroRNA regulation in infectious diseases and its potential as a biosensor in future aquaculture industry: a review. '''Molecules''' 28:4357. - [[SiouNing 2023 Molecules |»Bioblast link«]] | ||
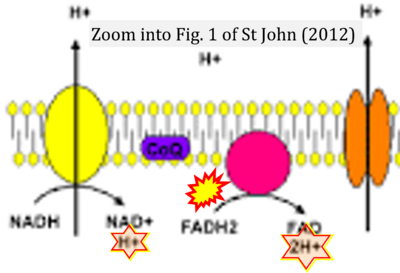
:::::: [[File: | :::::: [[File:St John 2012 Cell Tissue Res CORRECTION.png|400px|link=St John 2012 Cell Tissue Res]] | ||
:::: | :::: '''#16''' St John JC (2012) Transmission, inheritance and replication of mitochondrial DNA in mammals: implications for reproductive processes and infertility. '''Cell Tissue Res''' 349:795-808. - [[St John 2012 Cell Tissue Res |»Bioblast link«]] | ||
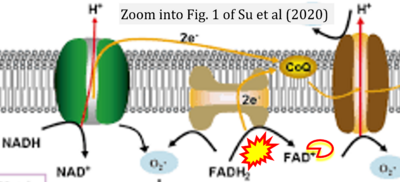
:::::: [[File: | :::::: [[File:Su 2020 Mol Biol Rep CORRECTION.png|400px|link=Su 2020 Mol Biol Rep]] | ||
:::: | :::: '''#17''' Su J, Ye D, Gao C, Huang Q, Gui D (2020) Mechanism of progression of diabetic kidney disease mediated by podocyte mitochondrial injury. '''Mol Biol Rep''' 47:8023-35. - [[Su 2020 Mol Biol Rep |»Bioblast link«]] | ||
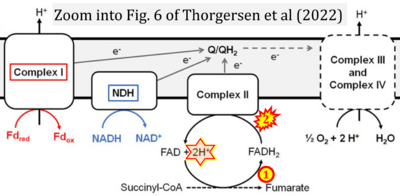
:::::: [[File: | :::::: [[File:Thorgersen 2022 Front Microbiol CORRECTION.png|400px|link=Thorgersen 2022 Front Microbiol]] | ||
:::: | :::: '''#18''' Thorgersen MP, Schut GJ, Poole FL 2nd, Haja DK, Putumbaka S, Mycroft HI, de Vries WJ, Adams MWW (2022) Obligately aerobic human gut microbe expresses an oxygen resistant tungsten-containing oxidoreductase for detoxifying gut aldehydes. '''Front Microbiol''' 13:965625. - [[Thorgersen 2022 Front Microbiol |»Bioblast link«]] | ||
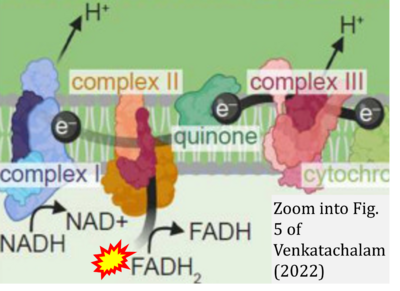
:::::: [[File: | :::::: [[File:Venkatachalam 2022 Cells CORRECTION.png|400px|link=Venkatachalam 2022 Cells]] | ||
:::: | :::: '''#19''' Venkatachalam K (2022) Regulation of aging and longevity by ion channels and transporters. '''Cells''' 11:1180. - [[Venkatachalam 2022 Cells |»Bioblast link«]] | ||
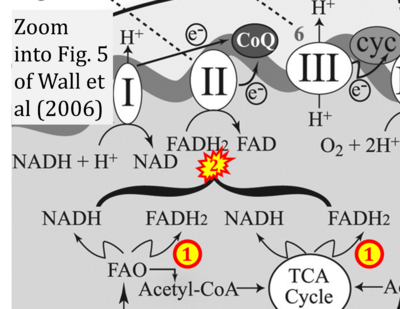
:::::: [[File: | :::::: [[File:Wall 2006 Am J Physiol Heart Circ Physiol CORRECTION.png|400px|link=Wall 2006 Am J Physiol Heart Circ Physiol]] | ||
:::: | :::: '''#20''' Wall JA, Wei J, Ly M, Belmont P, Martindale JJ, Tran D, Sun J, Chen WJ, Yu W, Oeller P, Briggs S, Gustafsson AB, Sayen MR, Gottlieb RA, Glembotski CC (2006) Alterations in oxidative phosphorylation complex proteins in the hearts of transgenic mice that overexpress the p38 MAP kinase activator, MAP kinase kinase 6. '''Am J Physiol Heart Circ Physiol''' 291:H2462-72. - [[Wall 2006 Am J Physiol Heart Circ Physiol |»Bioblast link«]] | ||
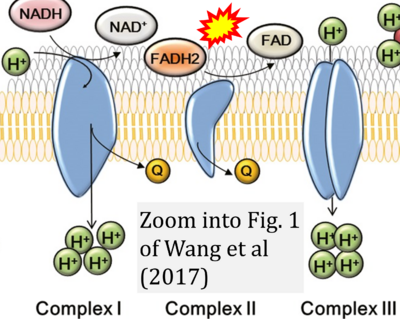
:::::: [[File: | :::::: [[File:Wang 2017 Am J Reprod Immunol CORRECTION.png|400px|link=Wang 2017 Am J Reprod Immunol]] | ||
:::: | :::: '''#21''' Wang T, Zhang M, Jiang Z, Seli E (2017) Mitochondrial dysfunction and ovarian aging. '''Am J Reprod Immunol''' 77. - [[Wang 2017 Am J Reprod Immunol |»Bioblast link«]] | ||
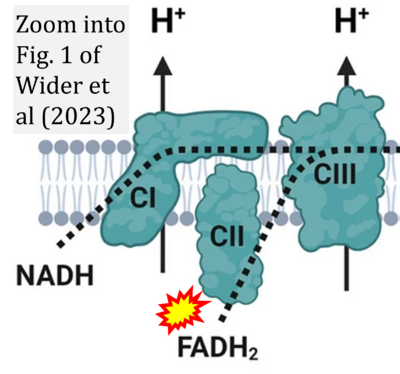
:::::: [[File: | :::::: [[File:Wider 2023 Crit Care CORRECTION.png|400px|link=Wider 2023 Crit Care]] | ||
:::: | :::: '''#22''' Wider JM, Gruley E, Morse PT, Wan J, Lee I, Anzell AR, Fogo GM, Mathieu J, Hish G, O’Neil B, Neumar RW, Przyklenk K, Hüttemann M, Sanderson TH (2023) Modulation of mitochondrial function with near-infrared light reduces brain injury in a translational model of cardiac arrest. '''Crit Care''' 27:491. - [[Wider 2023 Crit Care |»Bioblast link«]] | ||
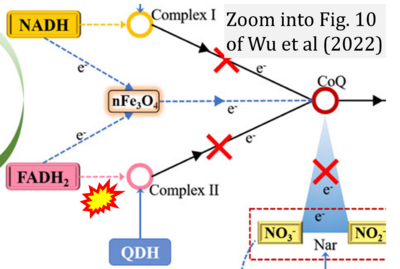
:::::: [[File: | :::::: [[File:Wu 2022 Front Chem CORRECTION.png|400px|link=Wu 2022 Front Chem]] | ||
:::: | :::: '''#23''' Wu Y, Liu X, Wang Q, Han D, Lin S (2022) Fe3O4-fused magnetic air stone prepared from wasted iron slag enhances denitrification in a biofilm reactor by increasing electron transfer flow. '''Front Chem''' 10:948453. - [[Wu 2022 Front Chem |»Bioblast link«]] | ||
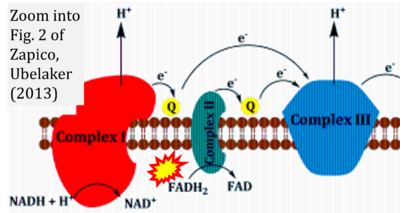
:::::: [[File: | :::::: [[File:Zapico 2013 Aging Dis CORRECTION.png|400px|link=Zapico 2013 Aging Dis]] | ||
:::: | :::: '''#24''' Zapico SC, Ubelaker DH (2013) mtDNA mutations and their role in aging, diseases and forensic sciences. '''Aging Dis''' 4:364-80. - [[Zapico 2013 Aging Dis |»Bioblast link«]] | ||
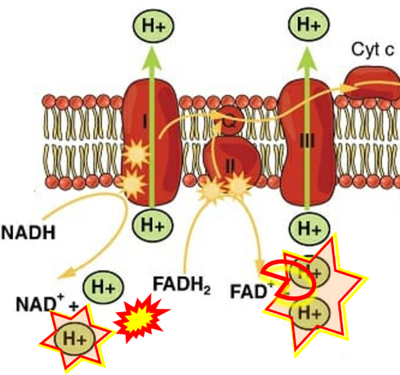
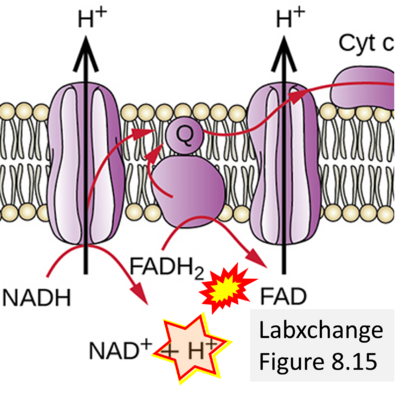
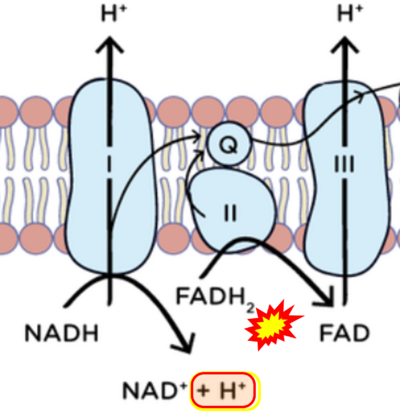
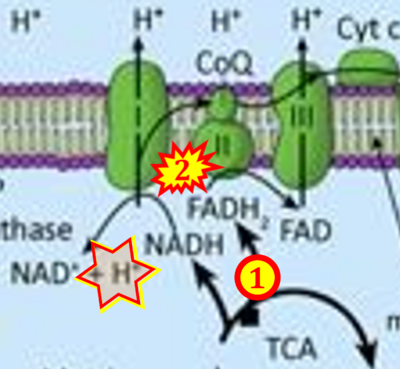
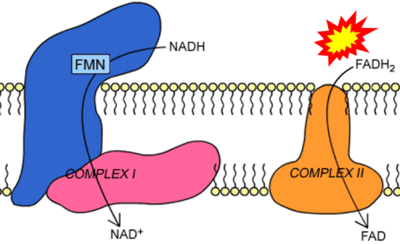
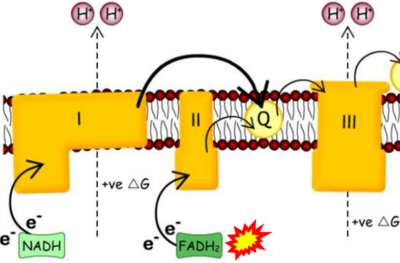
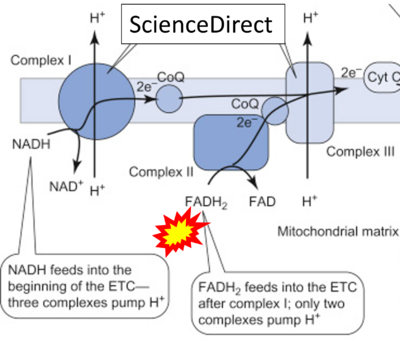
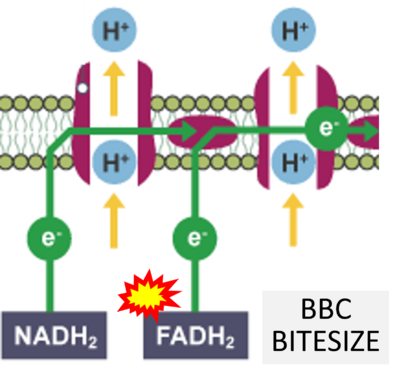
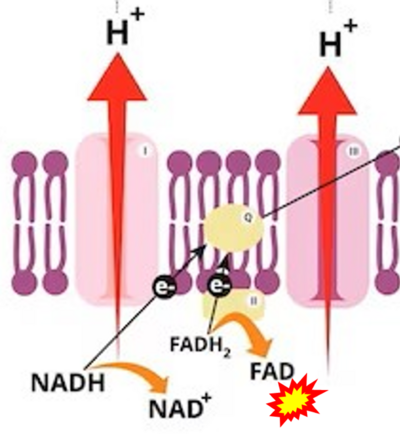
== | == Supplement: FADH<sub>2</sub> or FADH as substrate of CII in websites == | ||
:::: | :::: Complex II ambiguities in graphical representations on FADH<sub>2</sub> as a substrate of Complex II in the canonical forward electron transfer. FADH → FAD+H ('''g'''), FADH<sub>2</sub> → FAD+2H<sup>+</sup> ('''a’''', '''c''', '''h-n'''), and FADH<sub>2</sub> → FAD ('''a''', '''b''', '''d-f''', '''o-θ''') should be corrected to FADH<sub>2</sub> → FAD (Eq. 3b). NADH → NAD<sup>+</sup> is frequently written in graphs without showing the H<sup>+</sup> on the left side of the arrow, except for ('''p-r'''). NADH → NAD<sup>+</sup>+H<sup>+</sup> ('''a-g''', '''m'''), NADH → NAD<sup>+</sup>+2H<sup>+</sup> ('''h-l'''), NADH+H<sup>+</sup> → NAD<sup>+</sup>+2H<sup>+</sup> ('''j''', '''k'''), and NADH → NAD ('''ι''') should be corrected to NADH+H<sup>+</sup> → NAD<sup>+</sup> (Eq. 3a). (Retrieved 2023-03-21 to 2023-05-04). | ||
:::::: [[File:OpenStax Biology.png|400px]] | :::::: [[File:OpenStax Biology.png|400px]] | ||
:::: '''Website 1''': [https://openstax.org/books/biology/pages/7-4-oxidative-phosphorylation OpenStax Biology] - Fig. 7.10 Oxidative phosphorylation (CC BY 3.0). - OpenStax Biology got it wrong in figures and text. The error is copied without quality assessment and propagated in several links. | ::: ('''a''') | ||
:::: '''Website 2''': [https://opentextbc.ca/biology/chapter/4-3-citric-acid-cycle-and-oxidative-phosphorylation/ Concepts of Biology] - 1st Canadian Edition by Charles Molnar and Jane Gair - Fig. 4.19a | :::: '''Website 1''' ('''a''','''b'''): [https://openstax.org/books/biology/pages/7-4-oxidative-phosphorylation OpenStax Biology] - Fig. 7.10 Oxidative phosphorylation (CC BY 3.0). - OpenStax Biology got it wrong in figures and text. The error is copied without quality assessment and propagated in several links. | ||
:::: '''Website 3''': [https:// | :::: '''Website 2''' ('''a''','''b'''): [https://opentextbc.ca/biology/chapter/4-3-citric-acid-cycle-and-oxidative-phosphorylation/ Concepts of Biology] - 1st Canadian Edition by Charles Molnar and Jane Gair - Fig. 4.19a | ||
:::: '''Website | :::: '''Website 3''' ('''a''','''b'''): [https://www.pharmaguideline.com/2022/01/electron-transport-chain.html Pharmaguideline] | ||
:::: '''Website | :::: '''Website 4''' ('''a''','''b'''): [https://www.texasgateway.org/resource/74-oxidative-phosphorylation Texas Gateway] - Figure 7.11 | ||
:::: '''Website 5''' ('''a''','''b'''): [https://opened.cuny.edu/courseware/lesson/639/overview - CUNY] | |||
:::: '''Website 6''' ('''a''','''b'''): [https://courses.lumenlearning.com/wm-biology1/chapter/reading-electron-transport-chain/ lumen Biology for Majors I] - Fig. 1 | |||
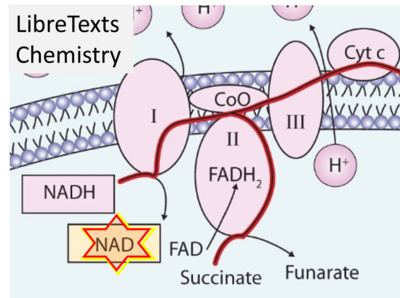
:::: '''Website 7''' ('''a'''): [https://bio.libretexts.org/Bookshelves/Introductory_and_General_Biology/Book%3A_General_Biology_(Boundless)/07%3A_Cellular_Respiration/7.11%3A_Oxidative_Phosphorylation_-_Electron_Transport_Chain LibreTexts Biology] Oxidative Phosphorylation - Electron Transport Chain - Figure 7.11.1 | |||
:::: '''Website 8''' ('''a'''): [https://brainbrooder.com/lesson/254/7-4-1-electron-transport-chain - Brain Brooder] | |||
:::::: [[File:Khan Academy modified from OpenStax CORRECTION.png| | :::::: [[File:Khan Academy modified from OpenStax CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''a’''') | ||
:::: '''Website | :::: '''Website 9''' ('''a’''','''b''','''v'''): [https://www.khanacademy.org/science/ap-biology/cellular-energetics/cellular-respiration-ap/a/oxidative-phosphorylation-etc Khan Academy] - Image modified from "Oxidative phosphorylation: Figure 1", by OpenStax College, Biology (CC BY 3.0). Figure and text underscore the FADH<sub>2</sub>-error: "''FADH<sub>2</sub> .. feeds them ''(electrons)'' into the transport chain through complex II.''" | ||
:::: '''Website 10''' ('''a’''','''b''','''v'''): [https://learn.saylor.org/mod/page/view.php?id=32815 Saylor Academy] | |||
:::::: [[File: | :::::: [[File:Expii OpenStax CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''b''') | ||
:::: '''Website 1''' ('''a''','''b'''): [https://openstax.org/books/biology/pages/7-4-oxidative-phosphorylation OpenStax Biology] - Fig. 7.12 | |||
:::: '''Website 2''' ('''a''','''b'''): [https://opentextbc.ca/biology/chapter/4-3-citric-acid-cycle-and-oxidative-phosphorylation/ Concepts of Biology] - 1st Canadian Edition by Charles Molnar and Jane Gair - Fig. 4.19c | |||
:::: '''Website 3''' ('''a''','''b'''): [https://www.pharmaguideline.com/2022/01/electron-transport-chain.html Pharmaguideline] | |||
:::: '''Website 4''' ('''a''','''b'''): [https://www.texasgateway.org/resource/74-oxidative-phosphorylation Texas Gateway] - Figure 7.13 | |||
:::: '''Website 5''' ('''a''','''b'''): [https://opened.cuny.edu/courseware/lesson/639/overview - CUNY] | |||
:::: '''Website 6''' ('''a''','''b'''): [https://courses.lumenlearning.com/wm-biology1/chapter/reading-electron-transport-chain/ lumen Biology for Majors I] - Fig. 3 | |||
:::: '''Website 9''' ('''a’''','''b''','''v'''): [https://www.khanacademy.org/science/ap-biology/cellular-energetics/cellular-respiration-ap/a/oxidative-phosphorylation-etc Khan Academy] - Image modified from "Oxidative phosphorylation: Figure 3," by Openstax College, Biology (CC BY 3.0) | |||
:::: '''Website 10''' ('''a’''','''b''','''v'''): [https://learn.saylor.org/mod/page/view.php?id=32815 Saylor Academy] | |||
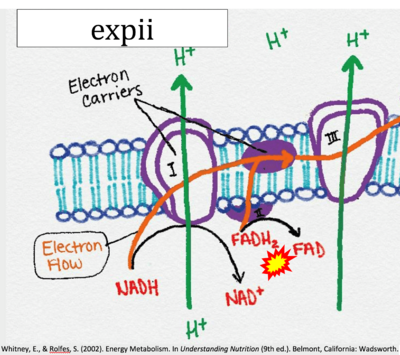
:::: '''Website 11''' ('''b''','''c''','''n''','''w''','''β'''): [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii] - Image source: By CNX OpenStax | |||
:::::: [[File: | :::::: [[File:Biologydictionary.net CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''c''') | ||
:::: '''Website | :::: '''Website 11''' ('''b''','''c''','''n''','''w''','''β'''): [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii] - Image source: By CNX OpenStax | ||
:::: '''Website | :::: '''Website 12''' ('''c''','''t'''): [https://www.thoughtco.com/electron-transport-chain-and-energy-production-4136143 ThoughtCo] - extender01 / iStock / Getty Images Plus | ||
:::: '''Website | :::: '''Website 13''' ('''c'''): [https://commons.wikimedia.org/w/index.php?curid=30148497 wikimedia 30148497 - Anatomy & Physiology, Connexions Web site. http://cnx.org/content/col11496/1.6/, 2013-06-19] | ||
:::: '''Website 14''' ('''c'''): [https://biologydictionary.net/electron-transport-chain-and-oxidative-phosphorylation/ biologydictionary.net 2018-08-21] | |||
:::: '''Website 15''' ('''c'''): [https://www.quora.com/Why-does-FADH2-form-2-ATP Quora] | |||
:::: '''Website 16''' ('''c'''): [https://teachmephysiology.com/biochemistry/atp-production/electron-transport-chain/ TeachMePhysiology] - Fig. 1. 2023-03-13 | |||
:::: '''Website 17''' ('''c'''): [https://www.toppr.com/ask/question/short-long-answer-types-whatis-the-electron-transport-system-and-what-are-its-functions/ toppr] | |||
:::::: [[File:Labxchange CORRECTION.png|400px]] | :::::: [[File:Labxchange CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''d''') | ||
:::: '''Website 18''' ('''d'''): [https://www.labxchange.org/library/items/lb:LabXchange:005ad47f-7556-3887-b4a6-66e74198fbcf:html:1 Labxchange] - Figure 8.15 credit: modification of work by Klaus Hoffmeier | |||
:::::: [[File:Jack Westin CORRECTION.png|400px]] | |||
::: ('''e''') | |||
:::: '''Website 19''' ('''e'''): [https://jackwestin.com/resources/mcat-content/oxidative-phosphorylation/electron-transfer-in-mitochondria Jack Westin MCAT Courses] | |||
:::::: [[File: | :::::: [[File:Videodelivery CORRECTION.png|400px]] | ||
::: | ::: ('''f''') | ||
:::: '''Website | :::: '''Website 20''' ('''f'''): [https://videodelivery.net/79e91c40bf96f9692560fa378c5086b6/thumbnails/thumbnail.jpg videodelivery] | ||
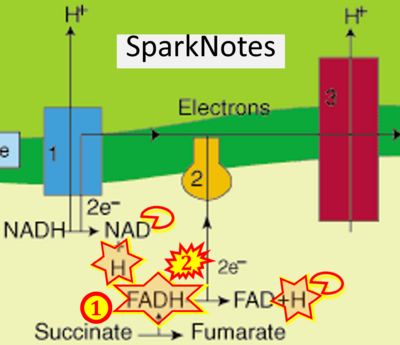
:::: | :::::: [[File:SparkNotes CORRECTION.png|400px]] | ||
::: | ::: ('''g''') | ||
:::: '''Website | :::: '''Website 21''' ('''g'''): [https://www.sparknotes.com/biology/cellrespiration/oxidativephosphorylation/section2/ - SparkNotes] | ||
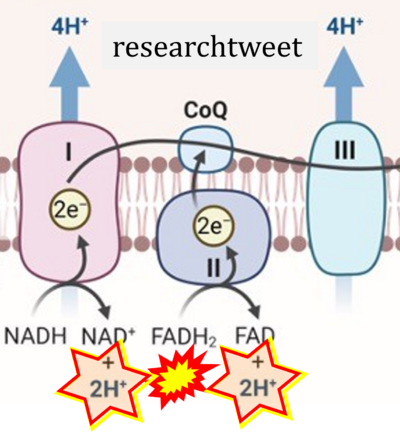
:::::: [[File:Researchtweet CORRECTION.png|400px]] | :::::: [[File:Researchtweet CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''h''') | ||
:::: '''Website | :::: '''Website 22''' ('''h''','''t'''): [https://researchtweet.com/mitochondrial-electron-transport-chain-2/ researchtweet] | ||
:::: '''Website 23''' ('''h'''): [https://microbenotes.com/electron-transport-chain/ Microbe Notes] | |||
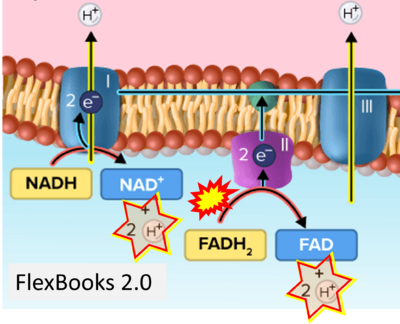
:::::: [[File: | :::::: [[File:FlexBooks 2 0 CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''i''') | ||
:::: '''Website 24''' ('''i'''): [https://flexbooks.ck12.org/cbook/ck-12-biology-flexbook-2.0/section/2.28/primary/lesson/electron-transport-bio/ FlexBooks] - CK-12 Biology for High School- 2.28 Electron Transport, Figure 2 | |||
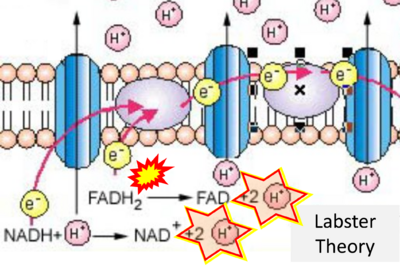
:::::: [[File:Labster Theory CORRECTION.png|400px]] | |||
::: ('''j''') | |||
:::: '''Website 25''' ('''j'''): [https://theory.labster.com/Electron_Transport_Chain/ Labster Theory] | |||
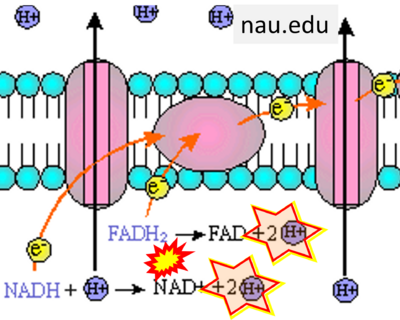
:::::: [[File:Nau.edu CORRECTION.png|400px]] | |||
::: ('''k''') | |||
:::: '''Website 26''' ('''k'''): [https://www2.nau.edu/~fpm/bio205/u4fg36.html nau.edu] | |||
:::::: [[File:ScienceFacts CORRECTION.png|400px]] | |||
::: ('''l''') | |||
:::: '''Website 27''' ('''l'''): [https://www.sciencefacts.net/electron-transport-chain.html ScienceFacts] | |||
:::::: [[File:Ck12 CORRECTION.png|400px]] | |||
::: ('''m''') | |||
:::: '''Website 28''' ('''m'''): [https://www.ck12.org/biology/electron-transport/lesson/The-Electron-Transport-Chain-Advanced-BIO-ADV/ cK-12] | |||
:::::: [[File:Wikimedia ETC CORRECTION.png|400px]] | |||
::: ('''n''') | |||
:::: '''Website 11''' ('''b''','''c''','''n''','''w''','''β'''): [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii] - Image source: By CNX OpenStax | |||
:::: '''Website 29''' ('''n'''): [https://commons.wikimedia.org/wiki/File:Mitochondrial_electron_transport_chain.png Wikimedia] | |||
:::::: [[File:Creative-biolabs CORRECTION.png|400px]] | |||
::: ('''o''') | |||
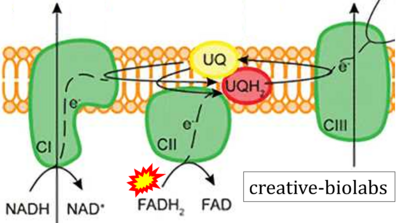
:::: '''Website 30''' ('''o'''): [https://www.creative-biolabs.com/drug-discovery/therapeutics/electron-transport-chain.htm creative-biolabs] | |||
:::::: [[File:Vector Mine CORRECTION.png|400px]] | :::::: [[File:Vector Mine CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''p''') | ||
:::: '''Website | :::: '''Website 31''' ('''p'''): [https://www.dreamstime.com/electron-transport-chain-as-respiratory-embedded-transporters-outline-diagram-electron-transport-chain-as-respiratory-embedded-image235345232 dreamstime] | ||
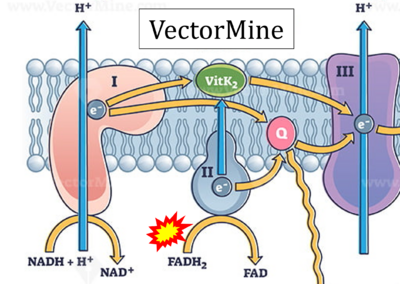
:::: '''Website 32''' ('''p'''): [https://vectormine.com/item/electron-transport-chain-as-respiratory-embedded-transporters-outline-diagram/ VectorMine] | |||
:::::: [[File: | :::::: [[File:YouTube Dirty Medicine Biochemistry CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''q''') | ||
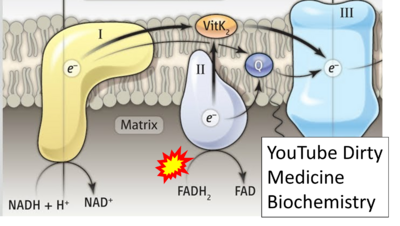
:::: '''Website 33''': [https://www.google.com/imgres?imgurl=https%3A%2F%2Fi.ytimg.com%2Fvi%2FLsRQ5_EmxJA%2Fmaxresdefault.jpg&tbnid=6w-0DVPMw7vOdM&vet=12ahUKEwjw2YO5--T9AhUwpCcCHduuDVgQMygDegUIARDzAQ..i&imgrefurl=https%3A%2F%2Fwww.youtube.com%2Fwatch%3Fv%3DLsRQ5_EmxJA&docid=bZxQYNch1Ys-VM&w=1280&h=720&q=electron%20transport%20chain&hl=en-US&client=firefox-b-d&ved=2ahUKEwjw2YO5--T9AhUwpCcCHduuDVgQMygDegUIARDzAQ YouTube Dirty Medicine Biochemistry] - Uploaded 2019-07-18 | |||
:::::: [[File: | :::::: [[File:DBriers CORRECTION.png|400px]] | ||
::: | ::: ('''r''') | ||
:::: '''Website | :::: '''Website 34''' ('''r'''): [http://www.dbriers.com/tutorials/ DBriers] | ||
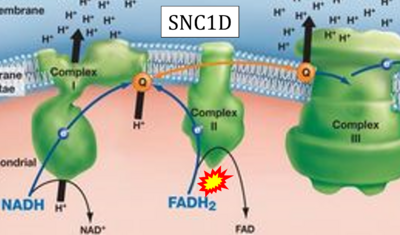
:::::: [[File: | :::::: [[File:SNC1D CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''s''') | ||
:::: '''Website 35''' ('''s'''): [https://sbi4uraft2014.weebly.com/electron-transport-chain.html SNC1D - BIOLOGY LESSON PLAN BLOG] | |||
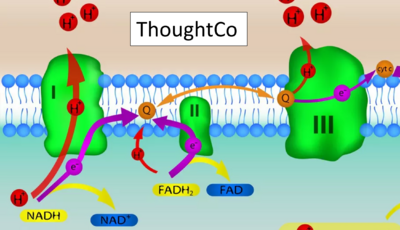
:::::: [[File: | :::::: [[File:ThoughtCo-Getty Images CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''t''') | ||
:::: '''Website 12''' ('''c''','''t'''): [https://www.thoughtco.com/electron-transport-chain-and-energy-production-4136143 ThoughtCo] - extender01 / iStock / Getty Images Plus | |||
:::: '''Website 22''' ('''h''','''t'''): [https://researchtweet.com/mitochondrial-electron-transport-chain-2/ researchtweet] | |||
:::: '''Website 36''' ('''t'''): [https://www.dreamstime.com/royalty-free-stock-photography-electron-transport-chain-illustration-oxidative-phosphorylation-image36048617 dreamstime] | |||
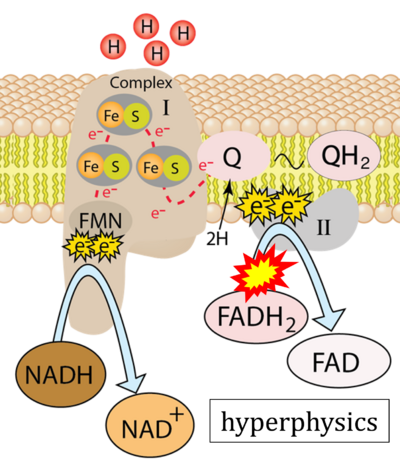
:::::: [[File:Hyperphysics CORRECTION.png|400px]] | :::::: [[File:Hyperphysics CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''u''') | ||
:::: '''Website 37''' ('''u'''): [http://hyperphysics.phy-astr.gsu.edu/hbase/Biology/Complex1.html hyperphysics] | |||
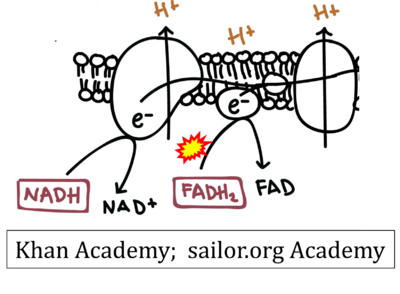
:::::: [[File:Khan Academy CORRECTION.png|400px]] | |||
::: ('''v''') | |||
:::: '''Website 9''' ('''a’''','''b''','''v'''): [https://www.khanacademy.org/science/ap-biology/cellular-energetics/cellular-respiration-ap/a/oxidative-phosphorylation-etc Khan Academy] | |||
:::: '''Website 10''' ('''a’''','''b''','''v'''): [https://learn.saylor.org/mod/page/view.php?id=32815 Saylor Academy] | |||
:::::: [[File: | :::::: [[File:Expii-Whitney, Rolfes 2002 CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''w''') | ||
:::: '''Website 11''' ('''b''','''c''','''n''','''w''','''β'''): [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii] - Whitney, Rolfes 2002 | |||
:::::: [[File: | :::::: [[File:UrbanPro CORRECTION.png|400px]] | ||
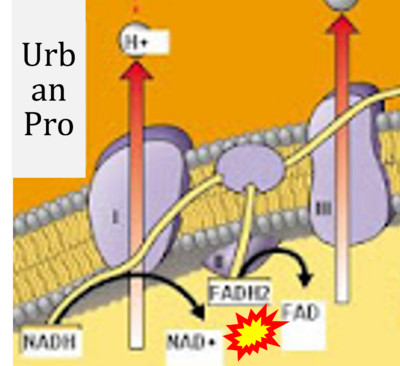
:::: '''Website | ::: ('''x''') | ||
:::: '''Website 38''' ('''x'''): [https://www.urbanpro.com/ba-tuition/oxidative-phosphorylation UrbanPro] | |||
:::::: [[File:Quizlet CORRECTION.png|400px]] | :::::: [[File:Quizlet CORRECTION.png|400px]] | ||
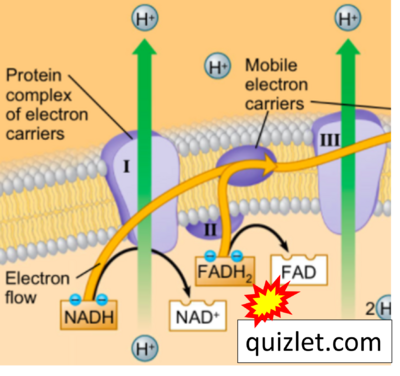
:::: '''Website | ::: ('''y''') | ||
:::: '''Website 39''' ('''y'''): [https://quizlet.com/245664214/electron-transport-chain-facts-of-cell-respiration-diagram/ Quizlet] | |||
:::::: [[File: | :::::: [[File:Unm.edu CORRECTION.png|400px]] | ||
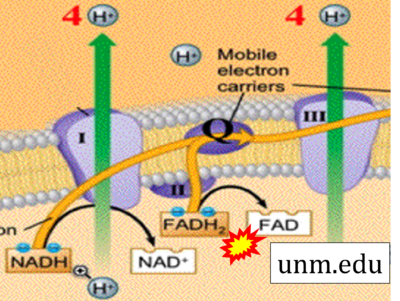
:::: '''Website | ::: ('''z''') | ||
:::: '''Website 40''' ('''z'''): [https://www.unm.edu/~lkravitz/Exercise%20Phys/ETCstory.html unm.edu] | |||
:::::: [[File:YouTube sciencemusicvideos CORRECTION.png|400px]] | |||
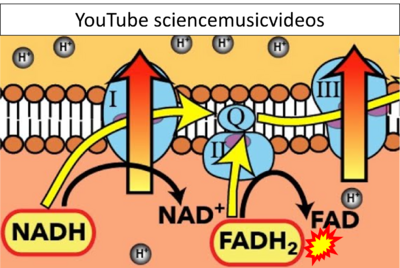
::: ('''α''') | |||
:::: '''Website 41''' ('''α'''): [https://www.google.com/imgres?imgurl=https%3A%2F%2Fi.ytimg.com%2Fvi%2FVER6xW_r1vc%2Fmaxresdefault.jpg&tbnid=Brshl0oN9LyYnM&vet=12ahUKEwjjlKSKpOX9AhWjmycCHbvGC34QMygWegUIARDWAQ..i&imgrefurl=https%3A%2F%2Fwww.youtube.com%2Fwatch%3Fv%3DVER6xW_r1vc&docid=VgTgrLf24Lzg4M&w=1280&h=720&itg=1&q=FADH2%20is%20the%20substrates%20of%20Complex%20II&hl=en&client=firefox-b-d&ved=2ahUKEwjjlKSKpOX9AhWjmycCHbvGC34QMygWegUIARDWAQ YouTube sciencemusicvideos] - Uploaded 2014-08-19 | |||
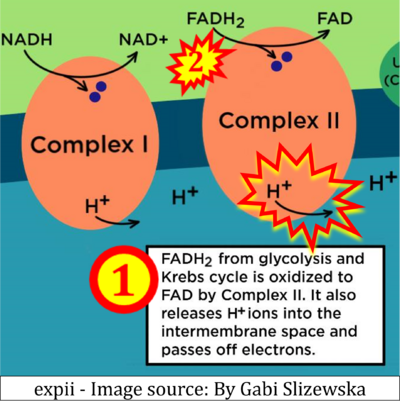
:::::: [[File:Expii-Gabi Slizewska CORRECTION.png|400px]] | |||
::: ('''β''') | |||
:::: '''Website 11''' ('''b''','''c''','''n''','''w''','''β'''): [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii expii] - Image source: By Gabi Slizewska | |||
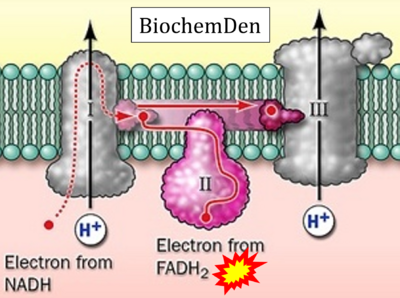
:::::: [[File: | :::::: [[File:BiochemDen CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''γ''') | ||
:::: '''Website 42''' ('''γ'''): [https://biochemden.com/electron-transport-chain-mechanism/ BiochemDen.com] | |||
:::::: [[File: | :::::: [[File:Hopes CORRECTION.png|400px]] | ||
:::: '''Website | :::('''δ''') | ||
:::: '''Website 43''' ('''δ'''): [https://hopes.stanford.edu/riboflavin/ hopes, Huntington’s outreach project for education, at Stanford] | |||
:::::: [[File: | :::::: [[File:Studocu CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''ε''') | ||
:::: '''Website 44''' ('''ε'''): [ https://www.studocu.com/en-gb/document/university-college-london/mammalian-physiology/electron-transport-chain/38063777 studocu, University College London] | |||
:::::: [[File: | :::::: [[File:ScienceDirect CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''ζ''') | ||
:::: '''Website 45''' ('''ζ'''): [https://www.google.com/imgres?imgurl=https%3A%2F%2Fars.els-cdn.com%2Fcontent%2Fimage%2F3-s2.0-B9780128008836000215-f21-07-9780128008836.jpg&imgrefurl=https%3A%2F%2Fwww.sciencedirect.com%2Ftopics%2Fengineering%2Felectron-transport-chain&tbnid=g3dD4u8Tvd6TWM&vet=12ahUKEwjc9deUprT9AhVxhv0HHXZbAd0QMygCegUIARDBAQ..i&docid=Moj_2_W0OpUDcM&w=632&h=439&q=FADH2%20is%20the%20substrates%20of%20Complex%20II&client=firefox-b-d&ved=2ahUKEwjc9deUprT9AhVxhv0HHXZbAd0QMygCegUIARDBAQ ScienceDirect] | |||
:::::: [[File: | :::::: [[File:BBC BITESIZE CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''η''') | ||
:::: '''Website 46''' ('''η'''): [https://www.bbc.co.uk/bitesize/guides/zdq9382/revision/5 BBC BITESIZE cK-12] | |||
:::::: [[File: | :::::: [[File:Freepik CORRECTION.png|400px]] | ||
:::: '''Website | ::: ('''θ''') | ||
:::: '''Website 47''' ('''θ'''): [https://www.freepik.com/premium-vector/oxidative-phosphorylation-process-electron-transport-chain-final-step-cellular-respiration_29211885.htm freepik] | |||
:::::: [[File: | :::::: [[File:LibreTexts Chemistry_CORRECTION.png|400px]] | ||
::: | ::: ('''ι''') | ||
:::: '''Website | :::: '''Website 48''' ('''ι'''): [https://chem.libretexts.org/Courses/Saint_Marys_College_Notre_Dame_IN/CHEM_118_(Under_Construction)/CHEM_118_Textbook/12%3A_Metabolism_(Biological_Energy)/12.4%3A_The_Citric_Acid_Cycle_and_Electron_Transport - LibreTexts Chemistry] - The Citric Acid Cycle and Electron Transport – Fig. 12.4.3 | ||
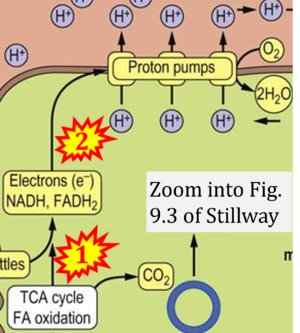
:::::: [[File:Stillway LW CORRECTION.png|300px]] | |||
:::: '''xx''' Stillway L William (2017) CHAPTER 9 Bioenergetics and Oxidative Metabolism. In: [https://doctorlib.info/medical/biochemistry/11.html Medical Biochemistry] | |||
<br> | |||
<big>'''from FAO and CII ambiguitiy to CII as a H<sup>+</sup> in websites'''</big> | |||
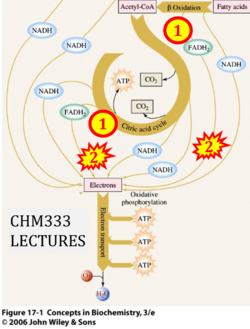
:::::: [[File: | :::::: [[File:CHM333 LECTURES CORRECTION.png|250px]] | ||
:::: | :::: '''xx''' [https://www.chem.purdue.edu/courses/chm333/Spring%202013/Lectures/Spring%202013%20Lecture%2037%20-%2038.pdf CHM333 LECTURES 37 & 38: 4/27 – 29/13 SPRING 2013 Professor Christine Hrycyna] | ||
<br> | |||
:::: '''Website | (retrieved 2023-03-21 to 2023-05-02) | ||
:::: '''Website 49''': [https://conductscience.com/electron-transport-chain/ Conduct Science]: "In Complex II, the enzyme succinate dehydrogenase in the inner mitochondrial membrane reduce FADH<sub>2</sub> to FAD<sup>+</sup>. Simultaneously, succinate, an intermediate in the Krebs cycle, is oxidized to fumarate." - Comments: FAD does not have a postive charge. FADH<sub>2</sub> is the reduced form, it is not reduced. And again: In CII, FAD is reduced to FADH<sub>2</sub>. | |||
:::: '''Website 50''': [https://themedicalbiochemistrypage.org/oxidative-phosphorylation-related-mitochondrial-functions/ The Medical Biochemistry Page]: ‘In addition to transferring electrons from the FADH<sub>2</sub> generated by SDH, complex II also accepts electrons from the FADH<sub>2</sub> generated during fatty acid oxidation via the fatty acyl-CoA dehydrogenases and from mitochondrial glycerol-3-phosphate dehydrogenase (GPD2) of the glycerol phosphate shuttle’ (Figure 8d). | |||
:::: '''Website | |||
:::: | :::: '''Website 51''': [https://www.chem.purdue.edu/courses/chm333/Spring%202013/Lectures/Spring%202013%20Lecture%2037%20-%2038.pdf CHM333 LECTURES 37 & 38: 4/27 – 29/13 SPRING 2013 Professor Christine Hrycyna]: Acyl-CoA dehydrogenase is listed under 'Electron transfer in Complex II'. | ||
:::: ''' | :::::: [[File:Expii-Gabi Slizewska CORRECTION.png|400px]] | ||
:::: '''xx''': [https://www.expii.com/t/electron-transport-chain-summary-diagrams-10139 expii expii - Image source: By Gabi Slizewska]: ‘FADH<sub>2</sub> from glycolysis and Krebs cycle is oxidized to FAD by Complex II. It also releases H<sup>+</sup> ions into the intermembrane space and passes off electrons’ (retrieved 2023-05-04). | |||
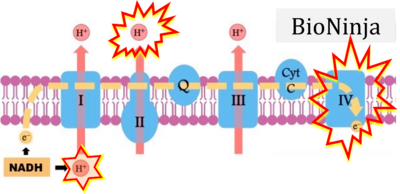
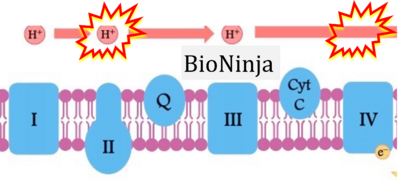
:::::: [[File:BioNinja 1 CORRECTION.png|400px]] | |||
:::::: [[File:BioNinja 2 CORRECTION.png|400px]] | |||
:::: '''xx''': [https://ib.bioninja.com.au/higher-level/topic-8-metabolism-cell/untitled/electron-transport-chain.html BioNinja] (retrieved 2023-05-04). | |||
{{Template:Keywords: Substrates and cofactors}} | |||
== Cited by == | |||
{{Template:Cited by Gnaiger 2024 MitoFit}} | |||
[[Category:Ambiguity crisis - CII and FADH2]] | |||
{{Labeling | {{Labeling | ||
|area= | |area=Patients, mt-Awareness | ||
| | |enzymes=Complex II;succinate dehydrogenase | ||
|additional=Ambiguity crisis, FAT4BRAIN, Publication:FAT4BRAIN | |||
|additional= | |||
}} | }} | ||
Latest revision as of 08:48, 1 May 2024
| Gnaiger E (2023) Complex II ambiguities ― FADH2 in the electron transfer system. MitoFit Preprints 2023.3.v6. https://doi.org/10.26124/mitofit:2023-0003.v6 - Published 2023-11-22 J Biol Chem (2024) |
» MitoFit Preprints 2023.3.v6.
Complex II ambiguities ― FADH2 in the electron transfer system
Gnaiger Erich (2023) MitoFit Prep
Abstract:
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470
- Version 6 (v6) 2023-06-21
- Version 5 (v5) 2023-05-31, (v4) 2023-05-12, (v3) 2023-05-04, (v2) 2023-04-04, (v1) 2023-03-24 - »Link to all versions«
The prevailing notion that reduced cofactors NADH and FADH2 transfer electrons from the tricarboxylic acid cycle to the mitochondrial electron transfer system creates ambiguities regarding respiratory Complex II (CII). The succinate dehydrogenase subunit SDHA of CII oxidizes succinate and reduces the covalently bound prosthetic group FAD to FADH2 in the canonical forward tricarboxylic acid cycle. However, several graphical representations of the electron transfer system depict FADH2 in the mitochondrial matrix as a substrate to be oxidized by CII. This leads to the false conclusion that FADH2 from the β-oxidation cycle in fatty acid oxidation feeds electrons into CII. In reality, dehydrogenases of fatty acid oxidation channel electrons to the coenzyme Q-junction but not through CII. The ambiguities surrounding Complex II in the literature and educational resources call for quality control, to secure scientific standards in current communications of bioenergetics, and ultimately support adequate clinical applications. This review aims to raise awareness of the inherent ambiguity crisis, complementing efforts to address the well-acknowledged issues of credibility and reproducibility.
• Keywords: coenzyme; cofactor; prosthetic group; coenzyme Q junction, Q-junction; Complex II, CII; H+-linked electron transfer; electron transfer system, ETS; matrix-ETS; membrane-ETS; fatty acid oxidation, FAO; flavin adenine dinucleotide, FAD/FADH2; nicotinamide adenine dinucleotide, NAD+/NADH; succinate dehydrogenase, SDH; tricarboxylic acid cycle, TCA; substrate; Gibbs force
• O2k-Network Lab: AT Innsbruck Oroboros
- » Links: Ambiguity crisis, Complex II ambiguities, Complex I and hydrogen ion ambiguities in the electron transfer system
- Acknowledgements: I thank Luiza H.D. Cardoso, Sabine Schmitt, and Chris Donnelly for stimulating discussions, and Paolo Cocco for expert help on the graphical abstract and Figures 1d and e. The constructive comments of an anonymous reviewer (J Biol Chem) are explicitly acknowledged. Contribution to the European Union’s Horizon 2020 research and innovation program Grant 857394 (FAT4BRAIN).
Additions to 312 references on CII-ambiguities after publication of JBC 2024
Last update 2023-12-19
- #1 Bektas A, Schurman SH, Gonzalez-Freire M, Dunn CA, Singh AK, Macian F, Cuervo AM, Sen R, Ferrucci L (2019) Age-associated changes in human CD4+ T cells point to mitochondrial dysfunction consequent to impaired autophagy. Aging (Albany NY) 11:9234-63. - »Bioblast link«
- #2 Ben-Shachar D (2009) The interplay between mitochondrial complex I, dopamine and Sp1 in schizophrenia. J Neural Transm (Vienna) 116:1383-96. - »Bioblast link«
- #3 Bon E, Maksimovich NY, Dremza IK (2022) Alendronate-induced nephropathy. J Clin Case Rep Stud 3. - »Bioblast link«
- #4 Elsaeed EM, Hamad A, Erfan OS, Elshahat M, Ebrahim F (2021) Role played by hippocampal apoptosis, autophagy and necroptosis in pathogenesis of diabetic cognitive dysfunction: a review of literature. Medicine Updates 6:41-63. - »Bioblast link«
- #5 Facucho-Oliveira JM, St John JC (2009) The relationship between pluripotency and mitochondrial DNA proliferation during early embryo development and embryonic stem cell differentiation. Stem Cell Rev Rep 5:140-58. - »Bioblast link«
- #6 Iqbal T, Welsby PJ, Howarth FC, Bidasee K, Adeghate E, Singh J (2014) Effects of diabetes-induced hyperglycemia in the heart: biochemical and structural slterations. In: Turan B, Dhalla N (eds) Diabetic cardiomyopathy. Advances in biochemistry in health and disease 9. Springer, New York. - »Bioblast link«
- #7 Keogh MJ, Chinnery PF (2015) Mitochondrial DNA mutations in neurodegeneration. Biochim Biophys Acta 1847:1401-11. - »Bioblast link«
- #8 Kunst C, Schmid S, Michalski M, Tümen D, Buttenschön J, Müller M, Gülow K (2023) The influence of gut microbiota on oxidative stress and the immune system. Biomedicines 11:1388. - »Bioblast link«
- #9 Lal MA (2018) Respiration. In: Bhatla SC, Lal MA (eds) Plant physiology, development and metabolism. Springer, Singapore:253-314. - »Bioblast link«
- #10 Lane RH, Tsirka AE, Gruetzmacher EM (2000) Uteroplacental insufficiency alters cerebral mitochondrial gene expression and DNA in fetal and juvenile rats. Pediatr Res 47:792-7. - »Bioblast link«
- #11 Palma FR, Gantner BN, Sakiyama MJ, Kayzuka C, Shukla S, Lacchini R, Cunniff B, Bonini MG (2023) ROS production by mitochondria: function or dysfunction? Oncogene. - »Bioblast link«
- #12 Quintard H, Fontaine E, Ichai C (2018) Energy metabolism: from the organ to the cell. In: Ichai, C., Quintard, H., Orban, JC. (eds) Metabolic Disorders and Critically Ill Patients. Springer, Cham. - »Bioblast link«
- #13 Reiss AB, Ahmed S, Dayaramani C, Glass AD, Gomolin IH, Pinkhasov A, Stecker MM, Wisniewski T, De Leon J (2022) The role of mitochondrial dysfunction in Alzheimer's disease: A potential pathway to treatment. Exp Gerontol 164:111828. - »Bioblast link«
- #14 Saghiv MS, Sagiv MS (2020) Metabolism. In: Basic Exercise Physiology. Springer, Cham. - »Bioblast link«
- #15 SiouNing AS, Seong TS, Kondo H, Bhassu S (2023) MicroRNA regulation in infectious diseases and its potential as a biosensor in future aquaculture industry: a review. Molecules 28:4357. - »Bioblast link«
- #16 St John JC (2012) Transmission, inheritance and replication of mitochondrial DNA in mammals: implications for reproductive processes and infertility. Cell Tissue Res 349:795-808. - »Bioblast link«
- #17 Su J, Ye D, Gao C, Huang Q, Gui D (2020) Mechanism of progression of diabetic kidney disease mediated by podocyte mitochondrial injury. Mol Biol Rep 47:8023-35. - »Bioblast link«
- #18 Thorgersen MP, Schut GJ, Poole FL 2nd, Haja DK, Putumbaka S, Mycroft HI, de Vries WJ, Adams MWW (2022) Obligately aerobic human gut microbe expresses an oxygen resistant tungsten-containing oxidoreductase for detoxifying gut aldehydes. Front Microbiol 13:965625. - »Bioblast link«
- #19 Venkatachalam K (2022) Regulation of aging and longevity by ion channels and transporters. Cells 11:1180. - »Bioblast link«
- #20 Wall JA, Wei J, Ly M, Belmont P, Martindale JJ, Tran D, Sun J, Chen WJ, Yu W, Oeller P, Briggs S, Gustafsson AB, Sayen MR, Gottlieb RA, Glembotski CC (2006) Alterations in oxidative phosphorylation complex proteins in the hearts of transgenic mice that overexpress the p38 MAP kinase activator, MAP kinase kinase 6. Am J Physiol Heart Circ Physiol 291:H2462-72. - »Bioblast link«
- #21 Wang T, Zhang M, Jiang Z, Seli E (2017) Mitochondrial dysfunction and ovarian aging. Am J Reprod Immunol 77. - »Bioblast link«
- #22 Wider JM, Gruley E, Morse PT, Wan J, Lee I, Anzell AR, Fogo GM, Mathieu J, Hish G, O’Neil B, Neumar RW, Przyklenk K, Hüttemann M, Sanderson TH (2023) Modulation of mitochondrial function with near-infrared light reduces brain injury in a translational model of cardiac arrest. Crit Care 27:491. - »Bioblast link«
- #23 Wu Y, Liu X, Wang Q, Han D, Lin S (2022) Fe3O4-fused magnetic air stone prepared from wasted iron slag enhances denitrification in a biofilm reactor by increasing electron transfer flow. Front Chem 10:948453. - »Bioblast link«
- #24 Zapico SC, Ubelaker DH (2013) mtDNA mutations and their role in aging, diseases and forensic sciences. Aging Dis 4:364-80. - »Bioblast link«
Supplement: FADH2 or FADH as substrate of CII in websites
- Complex II ambiguities in graphical representations on FADH2 as a substrate of Complex II in the canonical forward electron transfer. FADH → FAD+H (g), FADH2 → FAD+2H+ (a’, c, h-n), and FADH2 → FAD (a, b, d-f, o-θ) should be corrected to FADH2 → FAD (Eq. 3b). NADH → NAD+ is frequently written in graphs without showing the H+ on the left side of the arrow, except for (p-r). NADH → NAD++H+ (a-g, m), NADH → NAD++2H+ (h-l), NADH+H+ → NAD++2H+ (j, k), and NADH → NAD (ι) should be corrected to NADH+H+ → NAD+ (Eq. 3a). (Retrieved 2023-03-21 to 2023-05-04).
- (a)
- Website 1 (a,b): OpenStax Biology - Fig. 7.10 Oxidative phosphorylation (CC BY 3.0). - OpenStax Biology got it wrong in figures and text. The error is copied without quality assessment and propagated in several links.
- Website 2 (a,b): Concepts of Biology - 1st Canadian Edition by Charles Molnar and Jane Gair - Fig. 4.19a
- Website 3 (a,b): Pharmaguideline
- Website 4 (a,b): Texas Gateway - Figure 7.11
- Website 5 (a,b): - CUNY
- Website 6 (a,b): lumen Biology for Majors I - Fig. 1
- Website 7 (a): LibreTexts Biology Oxidative Phosphorylation - Electron Transport Chain - Figure 7.11.1
- Website 8 (a): - Brain Brooder
- (a’)
- Website 9 (a’,b,v): Khan Academy - Image modified from "Oxidative phosphorylation: Figure 1", by OpenStax College, Biology (CC BY 3.0). Figure and text underscore the FADH2-error: "FADH2 .. feeds them (electrons) into the transport chain through complex II."
- Website 10 (a’,b,v): Saylor Academy
- (b)
- Website 1 (a,b): OpenStax Biology - Fig. 7.12
- Website 2 (a,b): Concepts of Biology - 1st Canadian Edition by Charles Molnar and Jane Gair - Fig. 4.19c
- Website 3 (a,b): Pharmaguideline
- Website 4 (a,b): Texas Gateway - Figure 7.13
- Website 5 (a,b): - CUNY
- Website 6 (a,b): lumen Biology for Majors I - Fig. 3
- Website 9 (a’,b,v): Khan Academy - Image modified from "Oxidative phosphorylation: Figure 3," by Openstax College, Biology (CC BY 3.0)
- Website 10 (a’,b,v): Saylor Academy
- Website 11 (b,c,n,w,β): expii - Image source: By CNX OpenStax
- (c)
- Website 11 (b,c,n,w,β): expii - Image source: By CNX OpenStax
- Website 12 (c,t): ThoughtCo - extender01 / iStock / Getty Images Plus
- Website 13 (c): wikimedia 30148497 - Anatomy & Physiology, Connexions Web site. http://cnx.org/content/col11496/1.6/, 2013-06-19
- Website 14 (c): biologydictionary.net 2018-08-21
- Website 15 (c): Quora
- Website 16 (c): TeachMePhysiology - Fig. 1. 2023-03-13
- Website 17 (c): toppr
- (d)
- Website 18 (d): Labxchange - Figure 8.15 credit: modification of work by Klaus Hoffmeier
- (e)
- Website 19 (e): Jack Westin MCAT Courses
- (f)
- Website 20 (f): videodelivery
- (g)
- Website 21 (g): - SparkNotes
- (h)
- Website 22 (h,t): researchtweet
- Website 23 (h): Microbe Notes
- (i)
- Website 24 (i): FlexBooks - CK-12 Biology for High School- 2.28 Electron Transport, Figure 2
- (j)
- Website 25 (j): Labster Theory
- (k)
- Website 26 (k): nau.edu
- (l)
- Website 27 (l): ScienceFacts
- (m)
- Website 28 (m): cK-12
- (o)
- Website 30 (o): creative-biolabs
- (p)
- Website 31 (p): dreamstime
- Website 32 (p): VectorMine
- (q)
- Website 33: YouTube Dirty Medicine Biochemistry - Uploaded 2019-07-18
- (r)
- Website 34 (r): DBriers
- (s)
- Website 35 (s): SNC1D - BIOLOGY LESSON PLAN BLOG
- (t)
- Website 12 (c,t): ThoughtCo - extender01 / iStock / Getty Images Plus
- Website 22 (h,t): researchtweet
- Website 36 (t): dreamstime
- (u)
- Website 37 (u): hyperphysics
- (v)
- Website 9 (a’,b,v): Khan Academy
- Website 10 (a’,b,v): Saylor Academy
- (w)
- Website 11 (b,c,n,w,β): expii - Whitney, Rolfes 2002
- (x)
- Website 38 (x): UrbanPro
- (y)
- Website 39 (y): Quizlet
- (z)
- Website 40 (z): unm.edu
- (α)
- Website 41 (α): YouTube sciencemusicvideos - Uploaded 2014-08-19
- (β)
- Website 11 (b,c,n,w,β): expii expii - Image source: By Gabi Slizewska
- (γ)
- Website 42 (γ): BiochemDen.com
- (δ)
- Website 43 (δ): hopes, Huntington’s outreach project for education, at Stanford
- (ε)
- Website 44 (ε): [ https://www.studocu.com/en-gb/document/university-college-london/mammalian-physiology/electron-transport-chain/38063777 studocu, University College London]
- (ζ)
- Website 45 (ζ): ScienceDirect
- (η)
- Website 46 (η): BBC BITESIZE cK-12
- (θ)
- Website 47 (θ): freepik
- (ι)
- Website 48 (ι): - LibreTexts Chemistry - The Citric Acid Cycle and Electron Transport – Fig. 12.4.3
- xx Stillway L William (2017) CHAPTER 9 Bioenergetics and Oxidative Metabolism. In: Medical Biochemistry
from FAO and CII ambiguitiy to CII as a H+ in websites
(retrieved 2023-03-21 to 2023-05-02)
- Website 49: Conduct Science: "In Complex II, the enzyme succinate dehydrogenase in the inner mitochondrial membrane reduce FADH2 to FAD+. Simultaneously, succinate, an intermediate in the Krebs cycle, is oxidized to fumarate." - Comments: FAD does not have a postive charge. FADH2 is the reduced form, it is not reduced. And again: In CII, FAD is reduced to FADH2.
- Website 50: The Medical Biochemistry Page: ‘In addition to transferring electrons from the FADH2 generated by SDH, complex II also accepts electrons from the FADH2 generated during fatty acid oxidation via the fatty acyl-CoA dehydrogenases and from mitochondrial glycerol-3-phosphate dehydrogenase (GPD2) of the glycerol phosphate shuttle’ (Figure 8d).
- Website 51: CHM333 LECTURES 37 & 38: 4/27 – 29/13 SPRING 2013 Professor Christine Hrycyna: Acyl-CoA dehydrogenase is listed under 'Electron transfer in Complex II'.
- xx: expii expii - Image source: By Gabi Slizewska: ‘FADH2 from glycolysis and Krebs cycle is oxidized to FAD by Complex II. It also releases H+ ions into the intermembrane space and passes off electrons’ (retrieved 2023-05-04).
- xx: BioNinja (retrieved 2023-05-04).
- Bioblast links: Substrates and cofactors - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Cofactor
- » Cofactor
- » Coenzyme, cosubstrate
- » Nicotinamide adenine dinucleotide
- » Coenzyme Q2
- » Prosthetic group
- » Flavin adenine dinucleotide
- Cofactor
- Referennces
- » Gnaiger E (2023) Complex II ambiguities ― FADH2 in the electron transfer system. MitoFit Preprints 2023.3.v6. https://doi.org/10.26124/mitofit:2023-0003.v6
- Referennces
Cited by
- Gnaiger E (2024) Addressing the ambiguity crisis in bioenergetics and thermodynamics. MitoFit Preprints 2024.3. https://doi.org/10.26124/mitofit:2024-0003
Labels: MiParea: Patients, mt-Awareness
Enzyme: Complex II;succinate dehydrogenase
Ambiguity crisis, FAT4BRAIN, Publication:FAT4BRAIN