Gnaiger 2023 MitoFit CII
| Gnaiger E (2023) Complex II ambiguities ― FADH2 in the electron transfer system. MitoFit Preprints 2023.3.v6. https://doi.org/10.26124/mitofit:2023-0003.v6 |
» MitoFit Preprints 2023.3.v6.
Complex II ambiguities ― FADH2 in the electron transfer system
Gnaiger Erich (2023) MitoFit Prep
Abstract:
- Version 6 (v6) 2023-06-21 10.26124/mitofit:2023-0003.v6
- Version 5 (v5) 2023-05-31
- Version 4 (v4) 2023-05-12
- Version 3 (v3) 2023-05-04
- Version 2 (v2) 2023-04-04
- Version 1 (v1) 2023-03-24 - »Link to all versions«
The prevailing notion that reduced cofactors NADH and FADH2 transfer electrons from the tricarboxylic acid cycle to the mitochondrial electron transfer system creates ambiguities regarding respiratory Complex II (CII). The succinate dehydrogenase subunit SDHA of CII oxidizes succinate and reduces the covalently bound prosthetic group FAD to FADH2 in the canonical forward tricarboxylic acid cycle. However, several graphical representations of the electron transfer system depict FADH2 in the mitochondrial matrix as a substrate to be oxidized by CII. This leads to the false conclusion that FADH2 from the β-oxidation cycle in fatty acid oxidation feeds electrons into CII. In reality, dehydrogenases of fatty acid oxidation channel electrons to the coenzyme Q-junction but not through CII. The ambiguities surrounding Complex II in the literature and educational resources call for quality control, to secure scientific standards in current communications of bioenergetics, and ultimately support adequate clinical applications. This review aims to raise awareness of the inherent ambiguity crisis, complementing efforts to address the well-acknowledged issues of credibility and reproducibility.
• Keywords: coenzyme Q junction, Q-junction; Complex II, CII; electron transfer system, ETS; fatty acid oxidation, FAO; flavin adenine dinucleotide, FAD/FADH2; nicotinamide adenine dinucleotide, NAD+/NADH; succinate dehydrogenase, SDH; tricarboxylic acid cycle, TCA
• O2k-Network Lab: AT Innsbruck Oroboros
ORCID: ![]() Gnaiger Erich, Oroboros Instruments, Innsbruck, Austria
Gnaiger Erich, Oroboros Instruments, Innsbruck, Austria
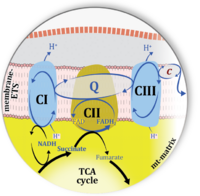
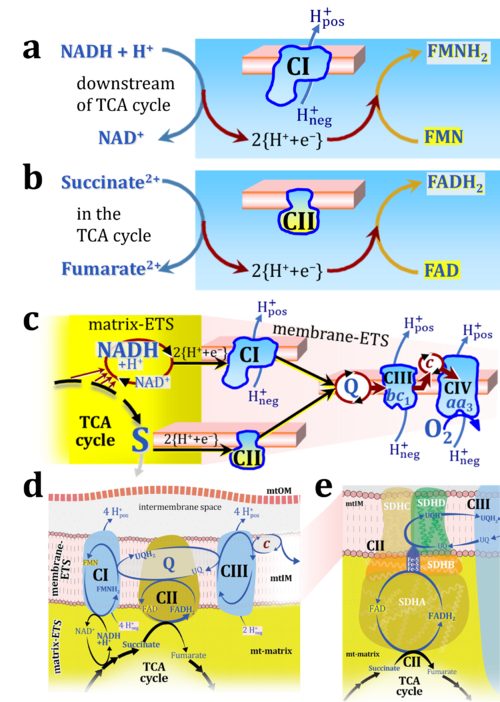
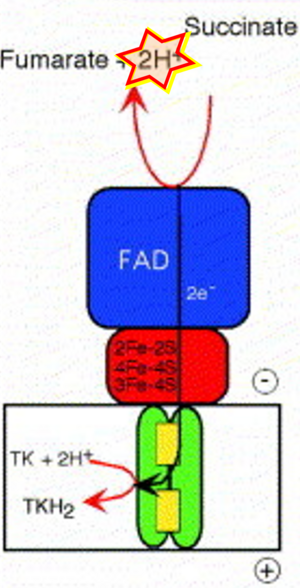
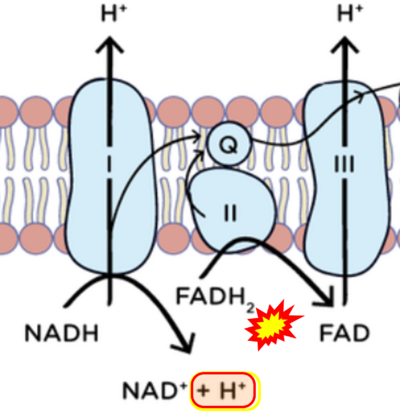
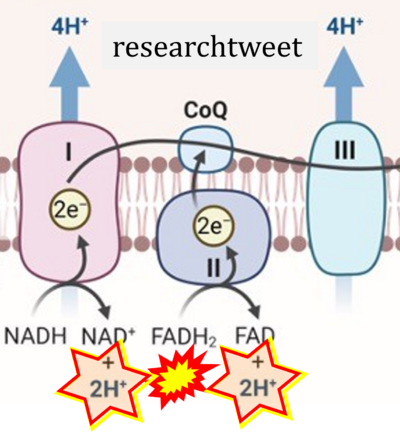
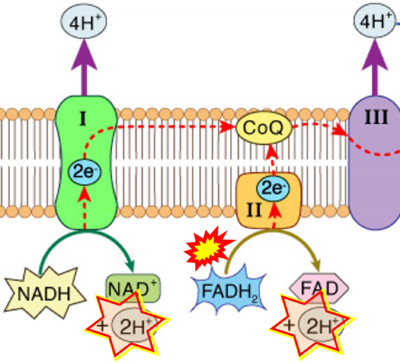
Figure 1. Complex II (SDH) bridges H+-linked electron transfer from the TCA cycle (matrix-ETS) to the electron transfer system (membrane-ETS) of the mt-inner membrane (mtIM). (a) NADH+H+ and (b) succinate are substrates of 2{H++e-} transfer to CI and CII, respectively, with prosthetic groups FMN and FAD as the corresponding electron acceptors. (c) Symbolic representation of ETS pathway architecture. Electron flow converges at the N-junction (NAD+ → NADH+H+). Electron flow from NADH and succinate S converges through CI and CII at the Q-junction. CIII passes electrons to cytochrome c and in CIV to molecular O2, 2{H++e-}+0.5 O2 ⇢ H2O. (d) NADH+H+ and NAD+ cycle between matrix-dehydrogenases and CI, whereas FAD and FADH2 cycle permanently bound within the same enzyme CII. Succinate and fumarate indicate the chemical entities irrespective of ionization, but charges are shown in NADH, NAD+, and H+. Joint pairs of half-circular arrows distinguish electron transfer 2{H++e-} to CI and CII from vectorial H+ translocation across the mtIM (H+neg → H+pos). CI and CIII pump hydrogen ions from the negatively (neg) to the positively charged compartment (pos). (e) Iconic representation of SDH subunits. SDHA catalyzes the oxidation succinate → fumarate + 2{H++e-} and reduction FAD + 2{H++e-} → FADH2 in the soluble domain of CII. The iron–sulfur protein SDHB transfers electrons through Fe-S clusters to the mtIM domain where ubiquinone UQ is reduced to ubiquinol UQH2 in SDHC and SDHD.
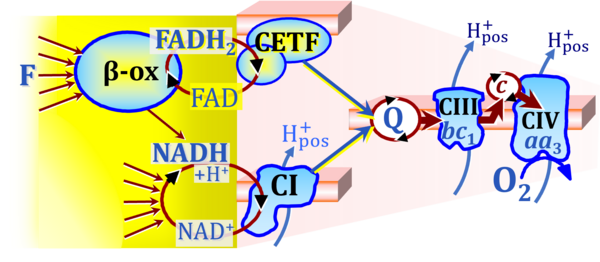
Figure 4. Fatty acid oxidation through the β-oxidation cycle (β-ox), the multi-enzyme electron transferring flavoprotein Complex (CETF, ETF:ETFDH; see text), and Complex I (CI) with convergent electron transfer into the Q-junction.
- Acknowledgements: I thank Luiza H. Cardoso and Sabine Schmitt for stimulating discussions, and Paolo Cocco for expert help on the graphical abstract and Figures 1d and e. The constructive comments of an anonymous reviewer (J Biol Chem) are explicitly acknowledged. Contribution to the European Union’s Horizon 2020 research and innovation program Grant 857394 (FAT4BRAIN).
Supplement 1. Footnotes on terminology
-
- ‘The dissociable, low-relative-molecular-mass active group of an enzyme which transfers chemical groups, hydrogen, or electrons. A coenzyme binds with its associated protein (apoenzyme) to form the active enzyme (holoenzyme) (Burtis, Geary 1994). ‘A low-molecular-weight, non-protein organic compound participating in enzymatic reactions as dissociable acceptor or donor of chemical groups or electrons’ - https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:23354 (CHEBI:23354, retrieved 2023-06-21). A coenzyme or cosubstrate is a cofactor that is attached loosely and transiently to an enzyme. NADH is listed as a coenzyme, which should be regarded as a substrate of pyridine-linked dehydrogenases (Lehninger 1975).
-
-
- A cofactor is 'an organic molecule or ion (usually a metal ion) that is required by an enzyme for its activity. It may be attached either loosely (coenzyme) or tightly (prosthetic group)' - https://www.ebi.ac.uk/chebi/searchId.do?chebiId=23357 (CHEBI:23357, retrieved 2023-06-21).
-
-
- The convergent architecture of the electron transfer system is emphasized in contrast to linear electron transfer chains ETCs within segments of the ETS.
-
- Electron transfer:
- A distinction is necessary between electron transfer in redox reactions and electron transport (translocation) in the diffusion of charged ionic species within or between cellular compartments. The symbol 2{H++e−} is introduced to indicate H+-linked electron transfer of two hydrogen ions and two electrons in a redox reaction.
- Gibbs force:
- In contrast to the extensive quantity Gibbs energy [J], Gibbs force [J·mol-1] is an intensive quantity expressed as the partial derivative of Gibbs energy [J] per advancement of a reaction [mol] (Gnaiger 1993; 2020).
- H+-linked electron transfer:
- The term H+-coupled electron transfer (Hsu et al 2022) is replaced by H+-linked electron transfer, to avoid confusion with coupled H+ translocation.
- Matrix-ETS:
- Electron transfer and corresponding OXPHOS capacities are classically studied in mitochondrial preparations as oxygen consumption supported by various fuel substrates undergoing partial oxidation in the mt-matrix, such as pyruvate, malate, succinate, and others. Therefore, the matrix component of ETS (matrix-ETS) is distinguished from the ETS bound to the mt-inner membrane (membrane-ETS; Gnaiger et al 2020).
- Membrane-ETS:
- Electron transfer is frequently considered as the segment of redox reactions linked to the mtIM. However, the membrane-ETS is only part of the total ETS, which includes the upstream matrix-ETS.
- Misinformation:
- Misinformation is the mistaken sharing of the same content (Wardle 2023).
-
- A prosthetic group is a cofactor that is ‘a tightly bound, specific nonpolypeptide unit in a protein determining and involved in its biological activity’ - https://www.ebi.ac.uk/chebi/searchId.do?chebiId=CHEBI:26348 (CHEBI:26348, retrieved 2023-06-21). A prosthetic group is attached permanently and tightly or even covalently to an enzyme and that is regenerated in each enzymatic turnover. FAD is the prosthetic group of flavin-linked dehydrogenases, covalently bound to CII.
-
-
- A substrate in a chemical reaction has a negative stoichiometric number since it is consumed, whereas a product has a positive stoichiometric number since it is produced. The general definition of a substrate in an enzyme-catalized reaction relies on the definition of the chemical reaction, without restriction to the nature of the substrate, i.e. independent of the substrate being a chemical entity in solution or a loosely bound cosubstrate (coenzyme) or even a tightly bound prosthetic group. The latter may be explicitly distinguished as a bound (internal) substrate from a free (external) substrate. Even different substrate pools may coexist (CoQ).
-
- 2{H++e-}
- In H+-linked two-electron transfer, 2H+ + 2e-, ‘the terms reducing equivalents or electron equivalents are used to refer to electrons and/or hydrogen atoms participating in oxidoreductions’ (Lehninger 1975). The symbol 2[H] is frequently used to distinguish reducing equivalents in the transfer from hydrogen donors to hydrogen acceptors from aqueous H+. Acid-base reactions obtain equilibrium fast without catalyst, whereas the slow oxidation-reduction reactions require an enzyme to proceed. However, 2[H] does not explicitly express that it applies to both electron and hydrogen ion transfer. Brackets are avoided to exclude the confusion with amount-of-substance concentrations frequently indicated by brackets. H+-linked two-electron transfer 2{H++e-} is distinguished from single-electron transfer {H+}+{e-}.
Beyond version 6
Last update: 2023-08-03
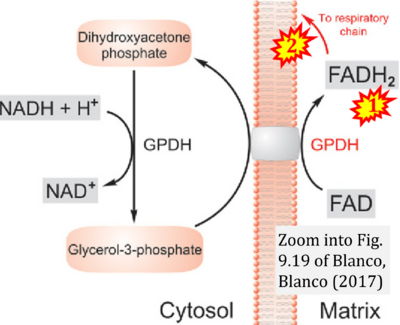
- From CGpDH to FADH2 to CII?
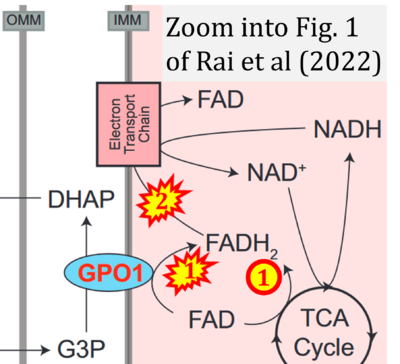
- Comment (Cardoso Luiza, Gnaiger Erich, 2023-08-03): Figure 9.19 (Blanco 2017 Academic Press) shows FADH2 to be formed in the mitochondrial matrix with a further arrow 'To respiratory chain'. Figure 1 (Rai 2022 G3 (Bethesda)) shows FADH2 to be formed in the mitochondrial matrix (1) from the TCA cycle feeding into the 'Electron transport chain' and (2) from GPO1 (CGpDH) feeding into the 'Electron transport chain'. Combined with FADH2 shown as a substrate of CII (on the right; one of >100 examples discussed as CII-ambiguities), one may arrive at the erroneous conclusion on a direct role of CII in the oxidation of glycerol-3-phosphate, analogous to false representations of CII involved in fatty acid oxidation.
Correction: FADH2 and Complex II
- FADH2 is shown as the substrate feeding electrons into Complex II (CII). This is wrong and requires correction - for details see Gnaiger (2024).
- Gnaiger E (2024) Complex II ambiguities ― FADH2 in the electron transfer system. J Biol Chem 300:105470. https://doi.org/10.1016/j.jbc.2023.105470 - »Bioblast link«
- Additions to Supplements
- Add to Supplement 2
- xx Pharaoh G, Kamat V, Kannan S, Stuppard RS, Whitson J, Martín-Pérez M, Qian WJ, MacCoss MJ, Villén J, Rabinovitch P, Campbell MD, Sweet IR, Marcinek DJ (2023) The mitochondrially targeted peptide elamipretide (SS-31) improves ADP sensitivity in aged mitochondria by increasing uptake through the adenine nucleotide translocator (ANT). Geroscience https://doi.org/10.1007/s11357-023-00861-y - »Bioblast link«
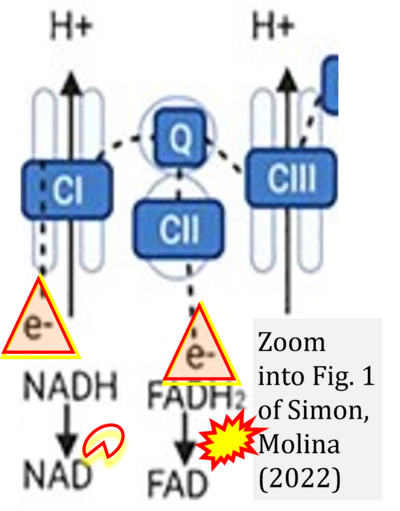
- xx Simon L, Molina PE (2022) Cellular bioenergetics: experimental evidence for alcohol-induced adaptations. Function (Oxf) 3:zqac039. - »Bioblast link«
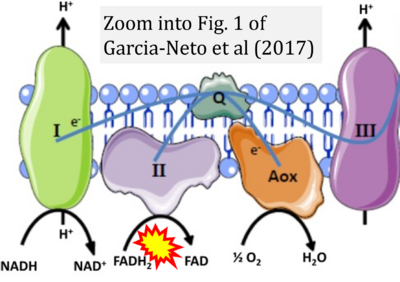
- xx Garcia-Neto W, Cabrera-Orefice A, Uribe-Carvajal S, Kowaltowski AJ, Alberto Luévano-Martínez L (2017) High osmolarity environments activate the mitochondrial alternative oxidase in Debaryomyces Hansenii. PLOS ONE 12:e0169621. - »Bioblast link«
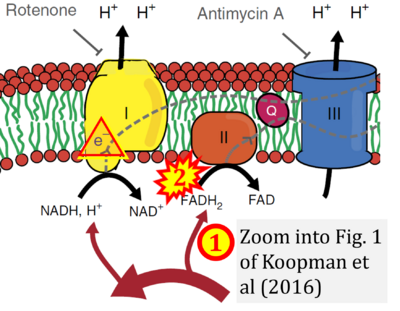
- xx Koopman M, Michels H, Dancy BM, Kamble R, Mouchiroud L, Auwerx J, Nollen EA, Houtkooper RH (2016) A screening-based platform for the assessment of cellular respiration in Caenorhabditis elegans. Nat Protoc 11:1798-816. - »Bioblast link«
- Add to Supplement 5
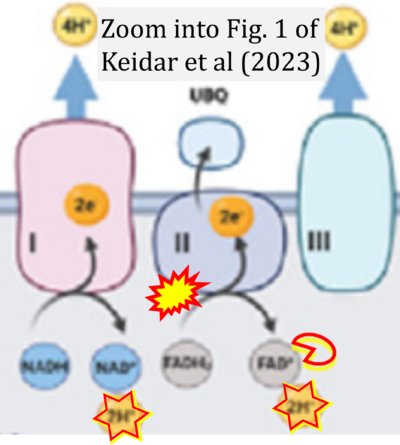
- xx Keidar N, Peretz NK and Yaniv Y (2023) Ca2+ pushes and pulls energetics to maintain ATP balance in atrial cells: computational insights. Front Physiol 14:1231259. - »Bioblast link«
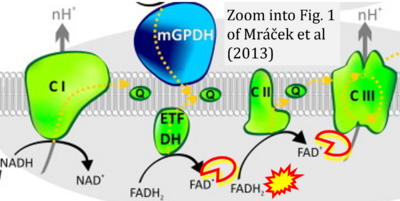
- xx Mracek T, Drahota Z, Houstek J (2013) The function and the role of the mitochondrial glycerol-3-phosphate dehydrogenase in mammalian tissues. Biochim Biophys Acta 1827:401-10. - »Bioblast link«
- Add to Supplement 6
- xx Diaz EC, Adams SH, Weber JL, Cotter M, Børsheim E (2023) Elevated LDL-C, high blood pressure, and low peak V˙O2 associate with platelet mitochondria function in children-The Arkansas Active Kids Study. Front Mol Biosci 10:1136975. - »Bioblast link«
- xx Steiner JL, Lang CH (2017) Etiology of alcoholic cardiomyopathy: Mitochondria, oxidative stress and apoptosis. Int J Biochem Cell Biol 89:125-35. - »Bioblast link«
- Beyond preprint
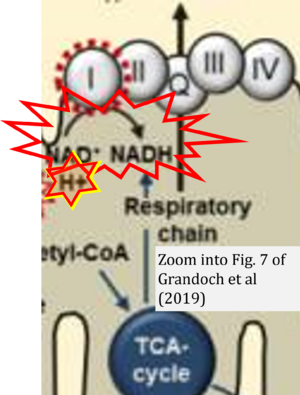
- 1 Grandoch M, Flögel U, Virtue S, Maier JK, Jelenik T, Kohlmorgen C, Feldmann K, Ostendorf Y, Castañeda TR, Zhou Z, Yamaguchi Y, Nascimento EBM, Sunkari VG, Goy C, Kinzig M, Sörgel F, Bollyky PL, Schrauwen P, Al-Hasani H, Roden M, Keipert S, Vidal-Puig A, Jastroch M5, Haendeler J, Fischer JW (2019) 4-Methylumbelliferone improves the thermogenic capacity of brown adipose tissue. Nat Metab 1:546-59. - »Bioblast link«
- NADH is shown as the product of the reaction catalyzed by CI in respiration. This error is rare in the literature, but comparable to the error frequenty encountered when FADH2 is shown as the substrate of CII.
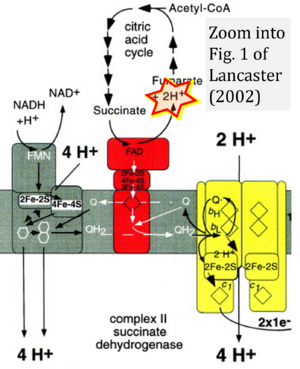
- 2 Lancaster CR (2002) Succinate:quinone oxidoreductases: an overview. Biochim Biophys Acta 1553:1-6. - »Bioblast link«
- fumarate + 2H+ shown besides NADH + H+ is ambiguous.
- 3 Lancaster CR (2001) Succinate:quinone oxidoreductases--what can we learn from Wolinella succinogenes quinol:fumarate reductase?. FEBS Lett 504:133-41. - »Bioblast link«
Supplement 2. FAD a substrate of SDH and FADH2 a substrate of CII
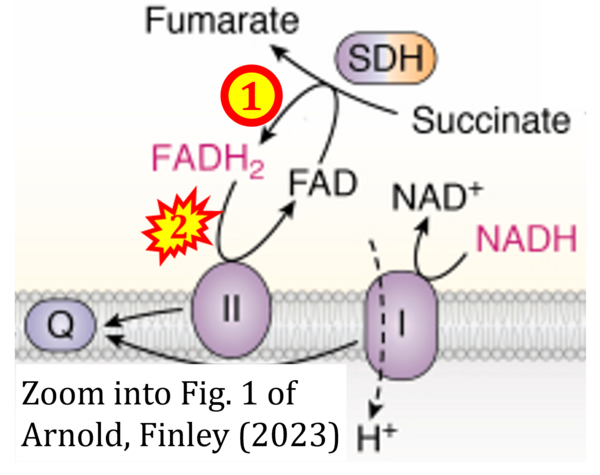
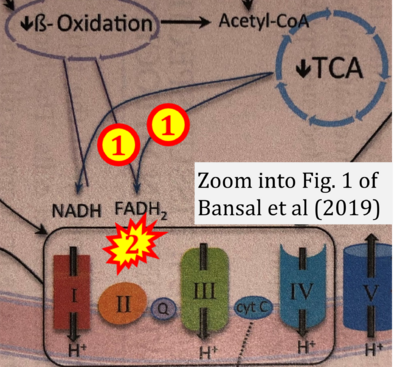
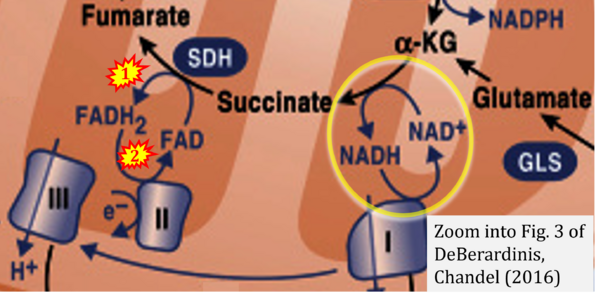
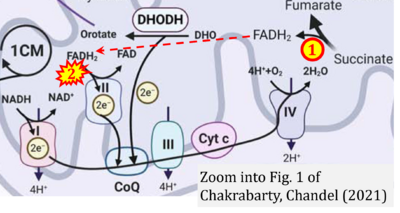
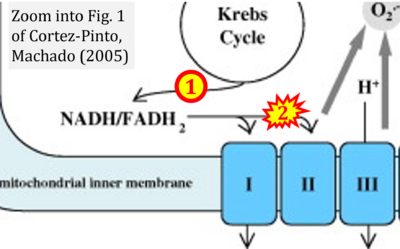
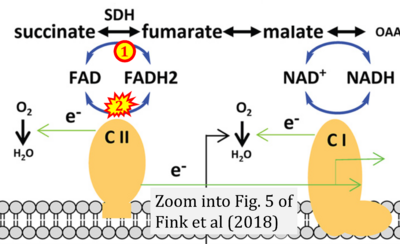
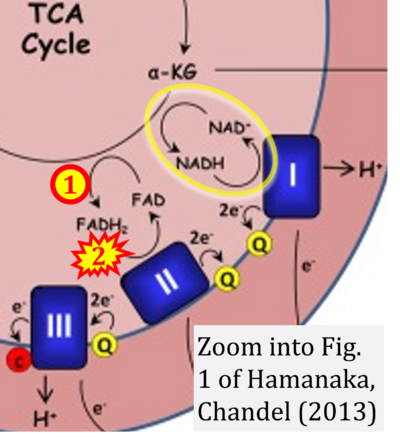
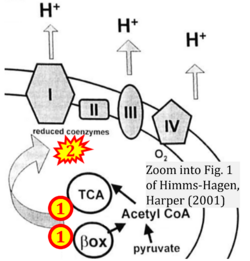
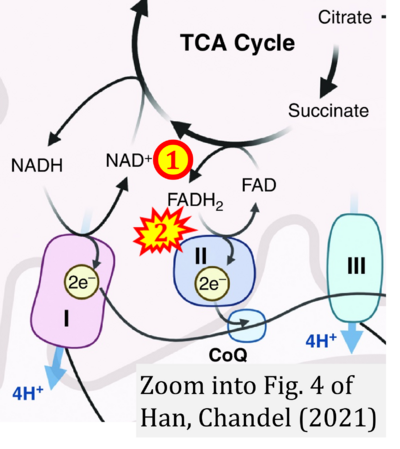
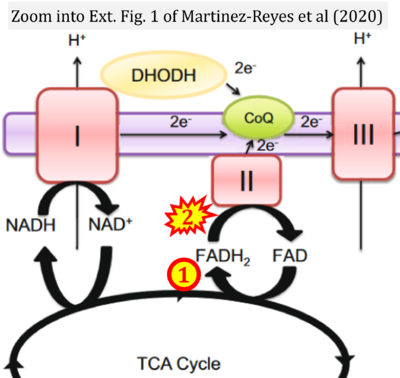
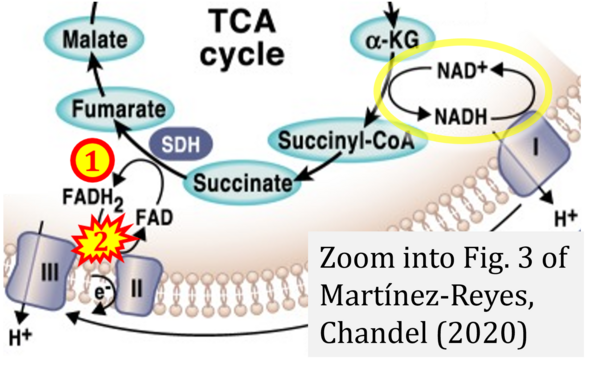
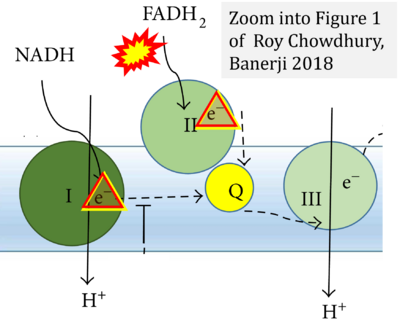
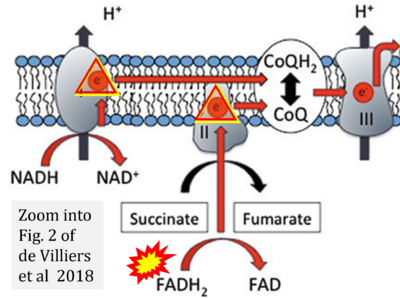
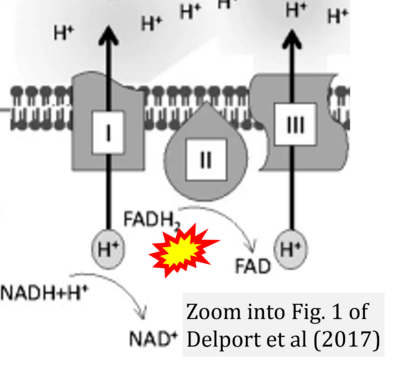
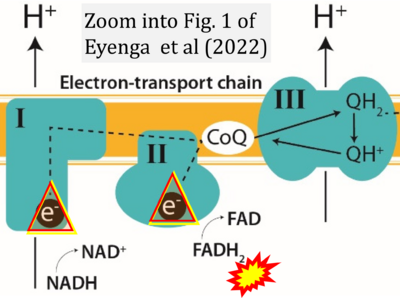
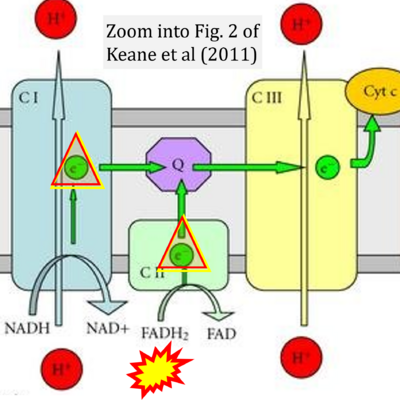
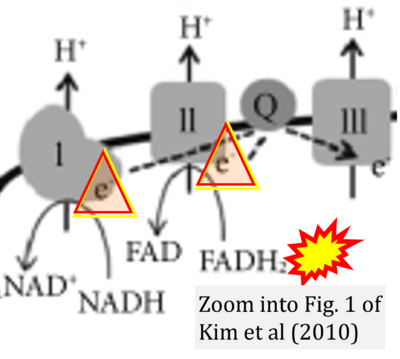
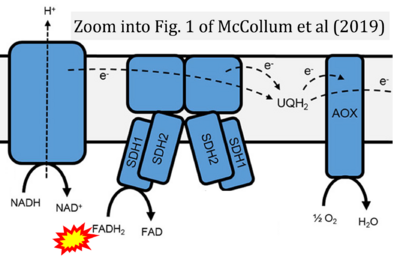
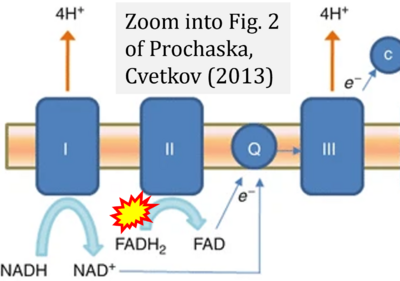
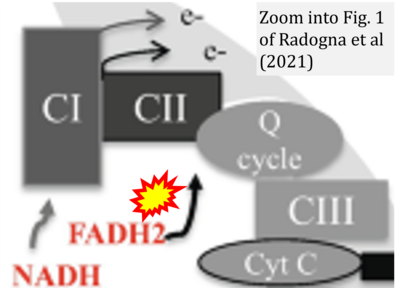
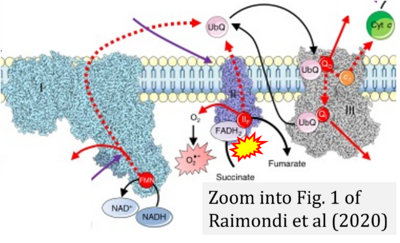
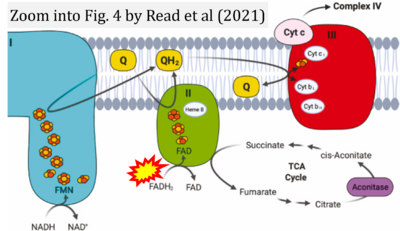
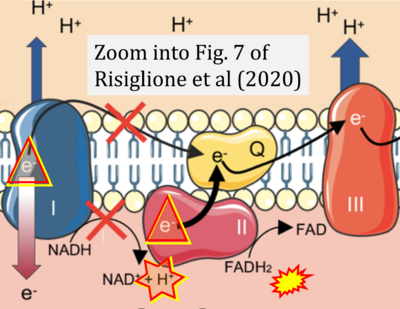
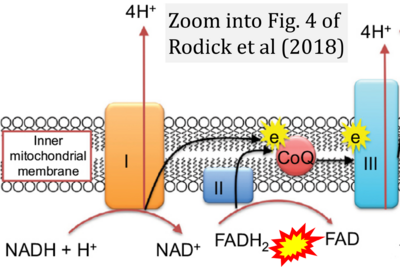
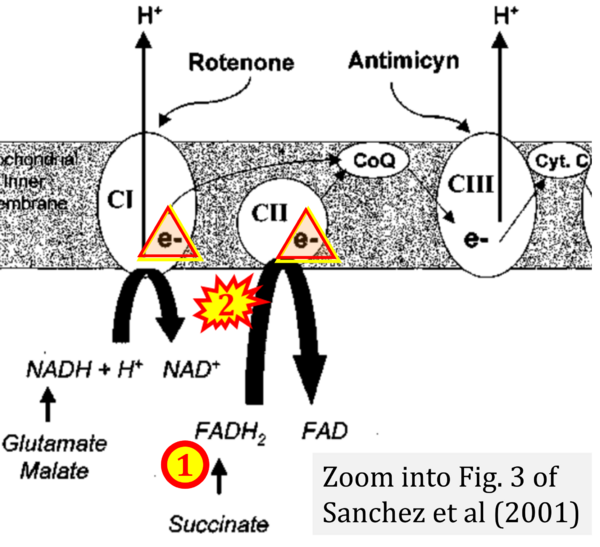
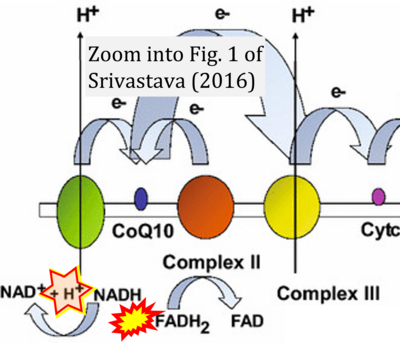
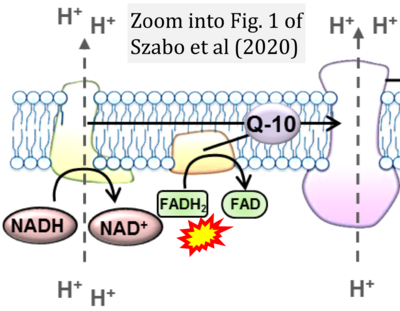
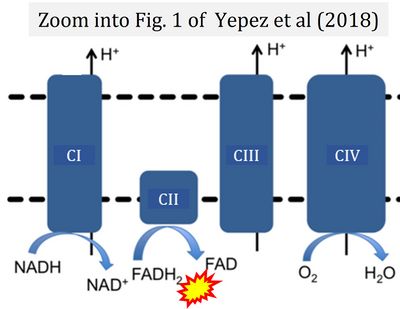
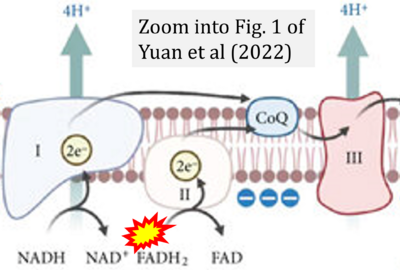
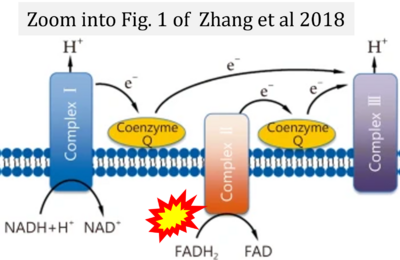
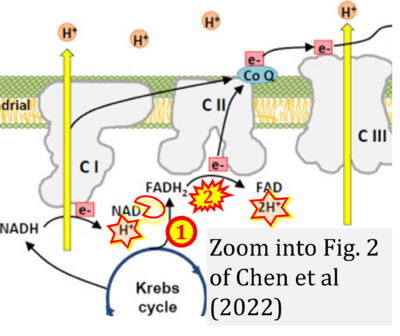
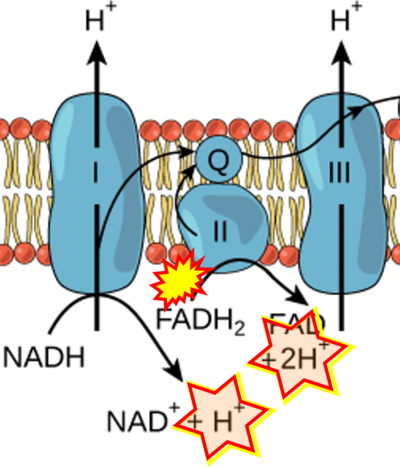
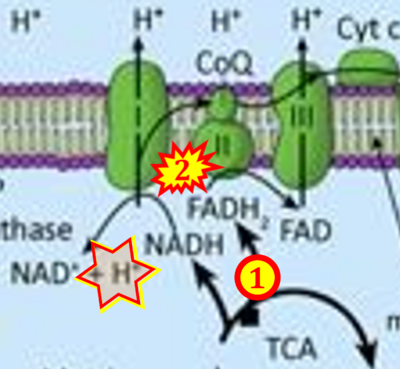
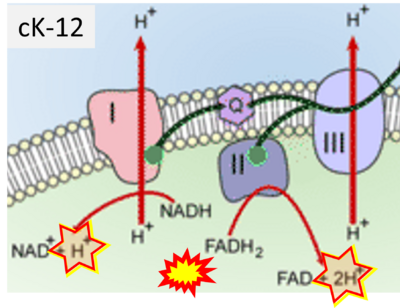
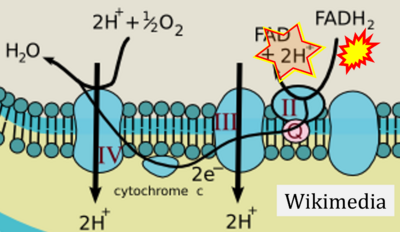
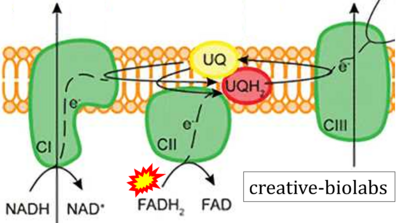
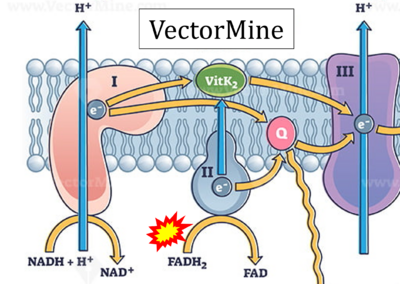
- Figure S2. Complex II ambiguities in graphical representations on FADH2 as a substrate of Complex II in the canonical forward electron transfer. The TCA cycle reduces FAD to FADH2 - in several cases shown to be catalyzed by SDH. Then FADH2 is erroneously shown to feed electrons into CII. Alphabetical sequence of publications from 2001 to 2023.
- a Arnold PK, Finley LWS (2023) Regulation and function of the mammalian tricarboxylic acid cycle. J Biol Chem 299:102838. - »Bioblast link«
- b Bansal A, Rashid C, Simmons RA (2019) Impact of fetal programming on mitochondrial function and susceptibility to obesity and type 2 diabetes. Academic Press In: Mitochondria in obesity and type 2 diabetes. Morio B, Pénicaud L, Rigoulet M (eds) Academic Press. - »Bioblast link«
- c Beier UH, Angelin A, Akimova T, Wang L, Liu Y, Xiao H, Koike MA, Hancock SA, Bhatti TR, Han R, Jiao J, Veasey SC, Sims CA, Baur JA, Wallace DC, Hancock WW (2015) Essential role of mitochondrial energy metabolism in Foxp3⁺ T-regulatory cell function and allograft survival. FASEB J 29:2315-26. - »Bioblast link«
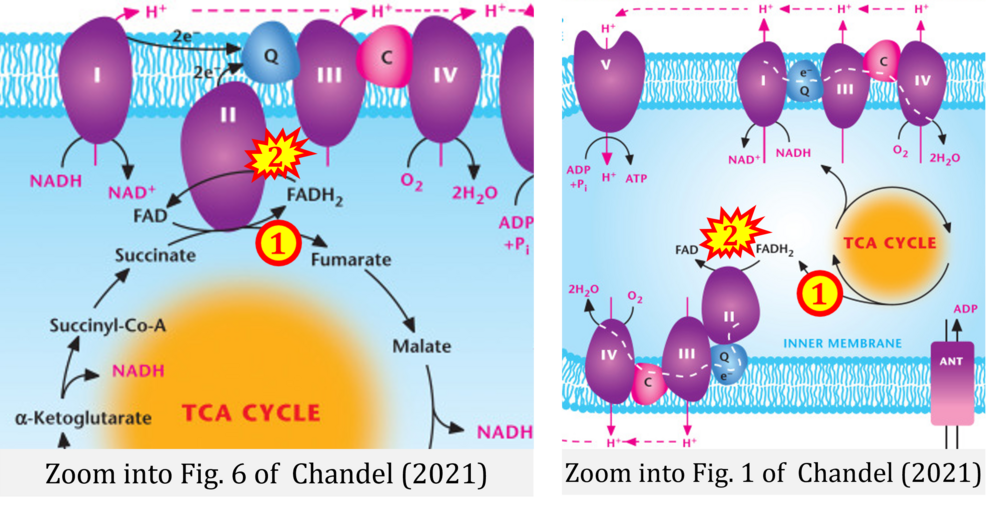
- d,e Chandel NS (2021) Mitochondria. Cold Spring Harb Perspect Biol 13:a040543. - »Bioblast link«
- f DeBerardinis RJ, Chandel NS (2016) Fundamentals of cancer metabolism. Sci Adv 2:e1600200. - »Bioblast link«
- g Chakrabarty RP, Chandel NS (2021) Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell 28:394-408. - »Bioblast link«
- h Cortez-Pinto H, Machado MV (2009) Uncoupling proteins and non-alcoholic fatty liver disease. J Hepatol 50:857-60. - »Bioblast link«
- i Fink BD, Bai F, Yu L, Sheldon RD, Sharma A, Taylor EB, Sivitz WI (2018) Oxaloacetic acid mediates ADP-dependent inhibition of mitochondrial complex II-driven respiration. J Biol Chem 293:19932-41. - »Bioblast link«
- j Hamanaka RB, Chandel NS (2013) Mitochondrial metabolism as a regulator of keratinocyte differentiation. Cell Logist 3:e25456. - »Bioblast link«
- k Himms-Hagen J, Harper ME (2001) Physiological role of UCP3 may be export of fatty acids from mitochondria when fatty acid oxidation predominates: an hypothesis. Exp Biol Med (Maywood) 226:78-84. - »Bioblast link«
- l Han S, Chandel NS (2021) Lessons from cancer metabolism for pulmonary arterial hypertension and fibrosis. Am J Respir Cell Mol Biol 65:134-45. - »Bioblast link«
- m Ishii I, Harada Y, Kasahara T (2012) Reprofiling a classical anthelmintic, pyrvinium pamoate, as an anti-cancer drug targeting mitochondrial respiration. Front Oncol 2:137. - »Bioblast link«
- n Jones PM, Bennett MJ (2017) Chapter 4 - Disorders of mitochondrial fatty acid β-oxidation. Elsevier In: Garg U, Smith LD , eds. Biomarkers in inborn errors of metabolism. Clinical aspects and laboratory determination:87-101. - »Bioblast link«
- o Lewis MT, Kasper JD, Bazil JN, Frisbee JC, Wiseman RW (2019) Quantification of mitochondrial oxidative phosphorylation in metabolic disease: application to Type 2 diabetes. Int J Mol Sci 20:5271. - »Bioblast link«
- p Martínez-Reyes I, Cardona LR, Kong H, Vasan K, McElroy GS, Werner M, Kihshen H, Reczek CR, Weinberg SE, Gao P, Steinert EM, Piseaux R, Budinger GRS, Chandel NS (2020) Mitochondrial ubiquinol oxidation is necessary for tumour growth. Nature 585:288-92. - »Bioblast link«
- q Martínez-Reyes I, Chandel NS (2020) Mitochondrial TCA cycle metabolites control physiology and disease. Nat Commun 11:102. - »Bioblast link«
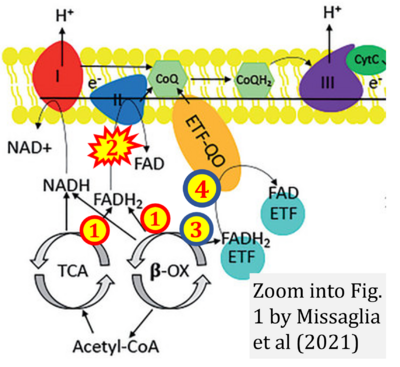
- r Missaglia S, Tavian D, Angelini C (2021) ETF dehydrogenase advances in molecular genetics and impact on treatment. Crit Rev Biochem Mol Biol 56:360-72. - »Bioblast link«
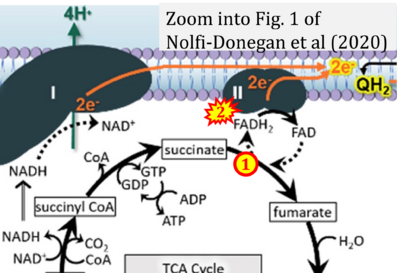
- s Nolfi-Donegan D, Braganza A, Shiva S (2020) Mitochondrial electron transport chain: Oxidative phosphorylation, oxidant production, and methods of measurement. Redox Biol 37:101674. - »Bioblast link«
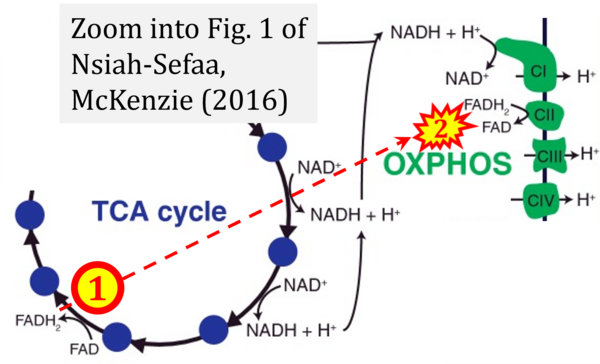
- t Nsiah-Sefaa A, McKenzie M (2016) Combined defects in oxidative phosphorylation and fatty acid β-oxidation in mitochondrial disease. Biosci Rep 36:e00313. - »Bioblast link«
- u Pelletier-Galarneau M, Detmer FJ, Petibon Y, Normandin M, Ma C, Alpert NM, El Fakhri G (2021) Quantification of myocardial mitochondrial membrane potential using PET. Curr Cardiol Rep 23:70. - »Bioblast link«
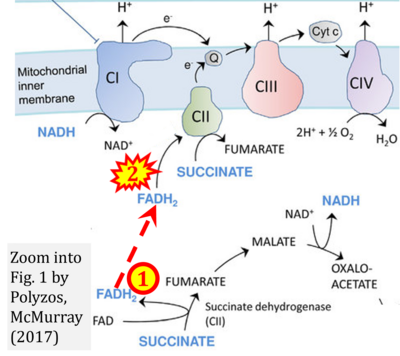
- v Polyzos AA, McMurray CT (2017) The chicken or the egg: mitochondrial dysfunction as a cause or consequence of toxicity in Huntington's disease. Mech Ageing Dev 161:181-97. - »Bioblast link«
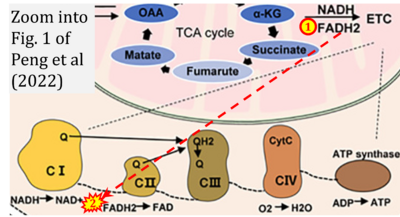
- w Peng M, Huang Y, Zhang L, Zhao X, Hou Y (2022) Targeting mitochondrial oxidative phosphorylation eradicates acute myeloid leukemic stem cells. Front Oncol 12:899502. - »Bioblast link«
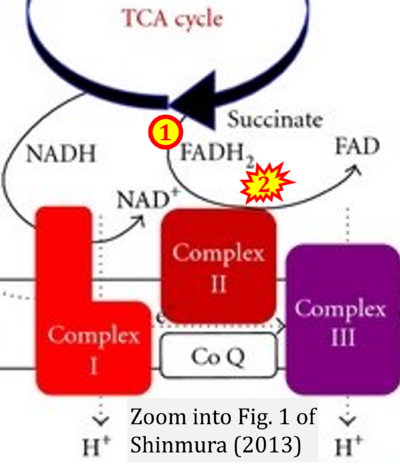
- x Shinmura K (2013) Effects of caloric restriction on cardiac oxidative stress and mitochondrial bioenergetics: potential role of cardiac sirtuins. Oxid Med Cell Longev 2013:528935. - »Bioblast link«
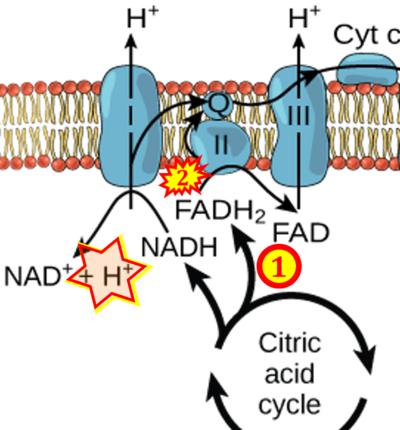
Supplement 3. FADH2 a substrate of CII
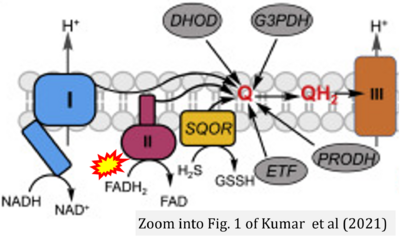
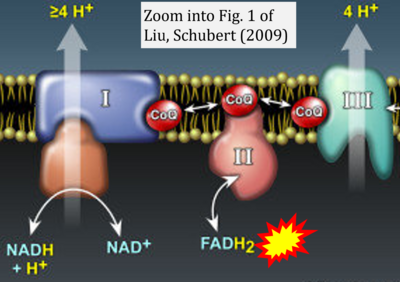
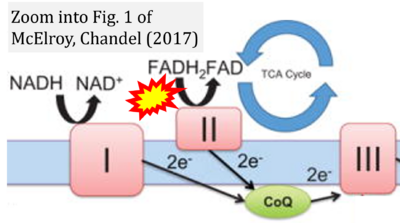
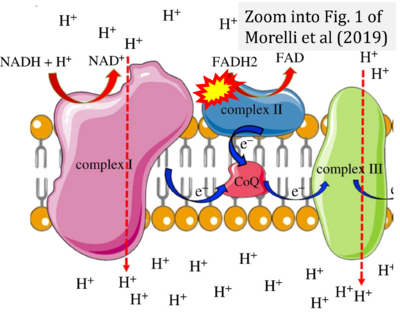
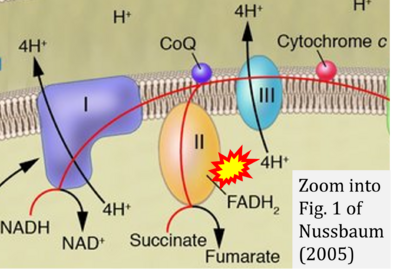
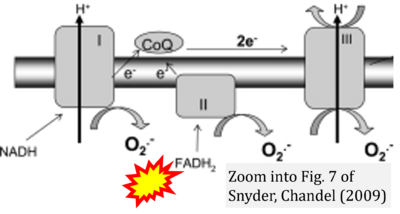
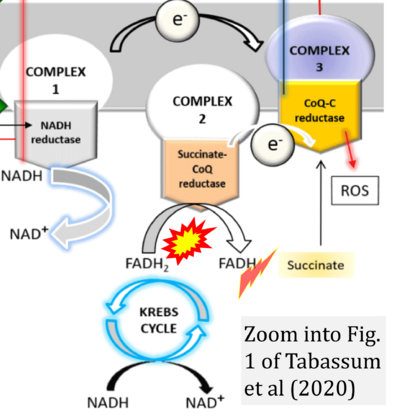
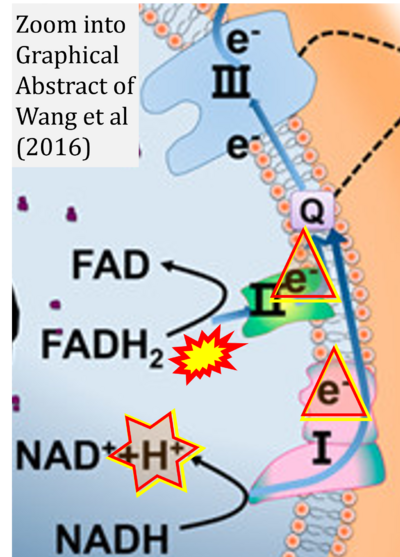
- Figure S3. Complex II ambiguities in graphical representations on FADH2 as a substrate of Complex II in the canonical forward electron transfer. Alphabetical sequence of publications from 2001 to 2023.
- a Balaban RS, Nemoto S, Finkel T (2005) Mitochondria, oxidants, and aging. Cell 120:483-95. - »Bioblast link«
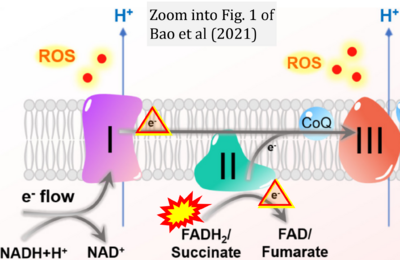
- b Bao MH, Wong CC (2021) Hypoxia, metabolic reprogramming, and drug resistance in liver cancer. Cells 10:1715. - »Bioblast link«
- c Benard G, Bellance N, Jose C, Rossignol R (2011) Relationships between mitochondrial dynamics and bioenergetics. In: Lu Bingwei (ed) Mitochondrial dynamics and neurodegeneration. Springer ISBN 978-94-007-1290-4:47-68. - »Bioblast link«
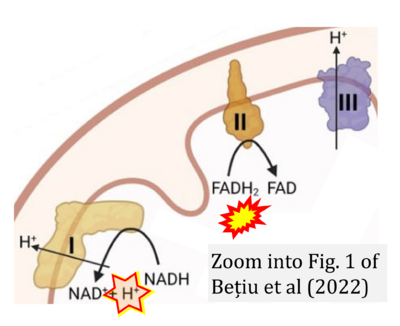
- d Bețiu AM, Noveanu L, Hâncu IM, Lascu A, Petrescu L, Maack C, Elmér E, Muntean DM (2022) Mitochondrial effects of common cardiovascular medications: the good, the bad and the mixed. Int J Mol Sci 23:13653. - »Bioblast link«
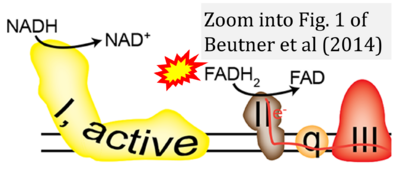
- e Beutner G, Eliseev RA, Porter GA Jr (2014) Initiation of electron transport chain activity in the embryonic heart coincides with the activation of mitochondrial complex 1 and the formation of supercomplexes. PLoS One 9:e113330. - »Bioblast link«
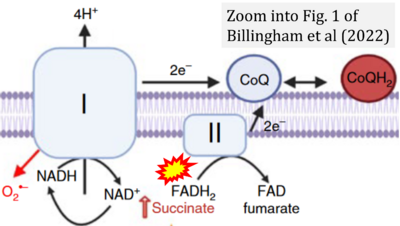
- f Billingham LK, Stoolman JS, Vasan K, Rodriguez AE, Poor TA, Szibor M, Jacobs HT, Reczek CR, Rashidi A, Zhang P, Miska J, Chandel NS (2022) Mitochondrial electron transport chain is necessary for NLRP3 inflammasome activation. Nat Immunol 23:692-704. - »Bioblast link«
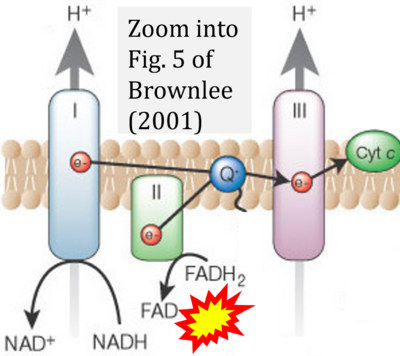
- g Brownlee M (2001) Biochemistry and molecular cell biology of diabetic complications. Nature 14:813-20. - »Bioblast link«
- Copied by: Arden GB, Ramsey DJ (2015) Diabetic retinopathy and a novel treatment based on the biophysics of rod photoreceptors and dark adaptation. In: Kolb H, Fernandez E, Nelson R, eds. Webvision: The organization of the retina and visual system [Internet]. Salt Lake City (UT): University of Utah Health Sciences Center; 1995-. - »Bioblast link«
- h Brownlee M (2003) A radical explanation for glucose-induced beta cell dysfunction. J Clin Invest 112:1788-90. - »Bioblast link«
- i Chakrabarty RP, Chandel NS (2021) Mitochondria as signaling organelles control mammalian stem cell fate. Cell Stem Cell 28:394-408. - »Bioblast link«
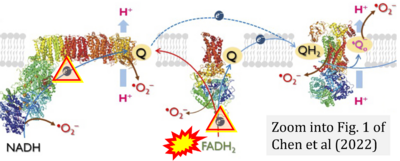
- j Chen CL, Zhang L, Jin Z, Kasumov T, Chen YR (2022) Mitochondrial redox regulation and myocardial ischemia-reperfusion injury. Am J Physiol Cell Physiol 322:C12-23. - »Bioblast link«
- k Roy Chowdhury S, Banerji V (2018) Targeting mitochondrial bioenergetics as a therapeutic strategy for chronic lymphocytic leukemia. Oxid Med Cell Longev 2018:2426712. - »Bioblast link«
- l de Beauchamp L, Himonas E, Helgason GV (2022) Mitochondrial metabolism as a potential therapeutic target in myeloid leukaemia. Leukemia 36:1-12. - »Bioblast link«
- m de Villiers D, Potgieter M, Ambele MA, Adam L, Durandt C, Pepper MS (2018) The role of reactive oxygen species in adipogenic differentiation. Adv Exp Med Biol 1083:125-144. - »Bioblast link«
- n Delport A, Harvey BH, Petzer A, Petzer JP (2017) Methylene blue and its analogues as antidepressant compounds. Metab Brain Dis 32:1357-82. - »Bioblast link«
- o Escoll P, Platon L, Buchrieser C (2019) Roles of mitochondrial respiratory Complexes during infection. Immunometabolism 1:e190011. - »Bioblast link«
- p Eyenga P, Rey B, Eyenga L, Sheu SS (2022) Regulation of oxidative phosphorylation of liver mitochondria in sepsis. Cells 11:1598. - »Bioblast link«
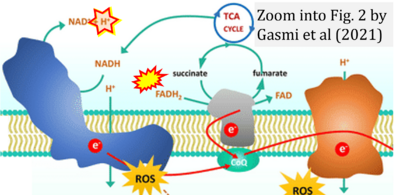
- q Gasmi A, Peana M, Arshad M, Butnariu M, Menzel A, Bjørklund G (2021) Krebs cycle: activators, inhibitors and their roles in the modulation of carcinogenesis. Arch Toxicol 95:1161-78. - »Bioblast link«
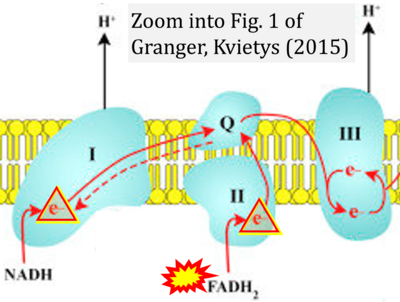
- r Granger DN, Kvietys PR (2015) Reperfusion injury and reactive oxygen species: The evolution of a concept. Redox Biol 6:524-551. - »Bioblast link«
- s Han S, Chandel NS (2019) There is no smoke without mitochondria. Am J Respir Cell Mol Biol 60:489-91. - »Bioblast link«
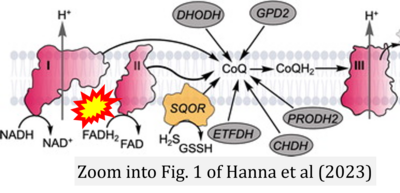
- t Hanna D, Kumar R, Banerjee R (2023) A metabolic paradigm for hydrogen sulfide signaling via electron transport chain plasticity. Antioxid Redox Signal 38:57-67. - »Bioblast link«
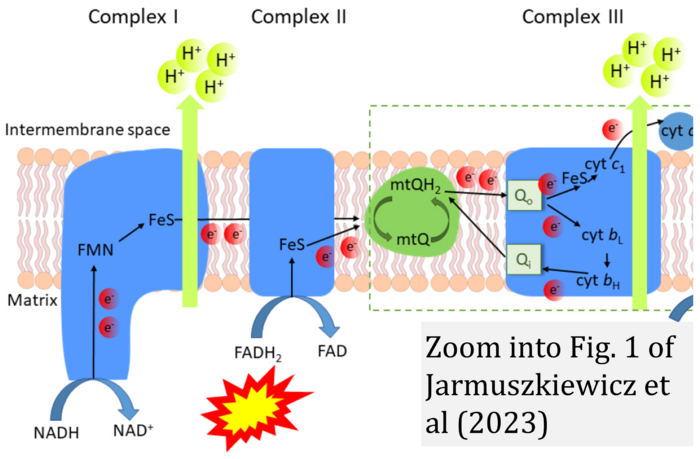
- u Jarmuszkiewicz W, Dominiak K, Budzinska A, Wojcicki K, Galganski L (2023) Mitochondrial coenzyme Q redox homeostasis and reactive oxygen species production. Front Biosci (Landmark Ed) 28:61. - »Bioblast link«
- v Keane PC, Kurzawa M, Blain PG, Morris CM (2011) Mitochondrial dysfunction in Parkinson's disease. Parkinsons Dis 2011:716871. - »Bioblast link«
- w Kim EH, Koh EH, Park JY, Lee KU (2010) Adenine nucleotide translocator as a regulator of mitochondrial function: implication in the pathogenesis of metabolic syndrome. Korean Diabetes J 34:146-53. - »Bioblast link«
- x Kumar R, Landry AP, Guha A, Vitvitsky V, Lee HJ, Seike K, Reddy P, Lyssiotis CA, Banerjee R (2021) A redox cycle with complex II prioritizes sulfide quinone oxidoreductase dependent H2S oxidation. J Biol Chem 298:101435. - »Bioblast link«
- y Liu Y, Schubert DR (2009) The specificity of neuroprotection by antioxidants. J Biomed Sci 16:98. - »Bioblast link«
- z Martínez-Reyes I, Diebold LP, Kong H, Schieber M, Huang H, Hensley CT, Mehta MM, Wang T, Santos JH, Woychik R, Dufour E, Spelbrink JN, Weinberg SE, Zhao Y, DeBerardinis RJ, Chandel NS (2016) TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol Cell 61:199-209. - »Bioblast link«
- α McCollum C, Geißelsöder S, Engelsdorf T, Voitsik AM, Voll LM (2019) Deficiencies in the mitochondrial electron transport chain affect redox poise and resistance toward Colletotrichum higginsianum. Front Plant Sci 10:1262. - »Bioblast link«
- β McElroy GS, Chandel NS (2017) Mitochondria control acute and chronic responses to hypoxia. Exp Cell Res 356:217-22. - »Bioblast link«
- γ McElroy GS, Reczek CR, Reyfman PA, Mithal DS, Horbinski CM, Chandel NS (2020) NAD+ regeneration rescues lifespan, but not ataxia, in a mouse model of brain mitochondrial Complex I dysfunction. Cell Metab 32:301-8.e6. - »Bioblast link«
- δ Morelli AM, Ravera S, Calzia D, Panfoli I (2019) An update of the chemiosmotic theory as suggested by possible proton currents inside the coupling membrane. Open Biol 9:180221. - »Bioblast link«
- ε Nussbaum RL (2005) Mining yeast in silico unearths a golden nugget for mitochondrial biology. J Clin Invest 115:2689-91. - »Bioblast link«
- ζ Prochaska LJ, Cvetkov TL (2013) Mitochondrial electron transport. In: Roberts, G.C.K. (eds) Encyclopedia of Biophysics. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-16712-6_25 - »Bioblast link«
- η Radogna F, Gerard D, Dicato M, Diederich M (2021) Assessment of mitochondrial cell metabolism by respiratory chain electron flow assays. Methods Mol Biol 2276:129-141. - »Bioblast link«
- θ Raimondi V, Ciccarese F, Ciminale V (2020) Oncogenic pathways and the electron transport chain: a dangeROS liaison. Br J Cancer 122:168-81. - »Bioblast link«
- ι Read AD, Bentley RE, Archer SL, Dunham-Snary KJ (2021) Mitochondrial iron-sulfur clusters: Structure, function, and an emerging role in vascular biology. Redox Biol 47:102164. - »Bioblast link«
- κ Risiglione P, Leggio L, Cubisino SAM, Reina S, Paternò G, Marchetti B, Magrì A, Iraci N, Messina A (2020) High-resolution respirometry reveals MPP+ mitochondrial toxicity mechanism in a cellular model of parkinson's disease. Int J Mol Sci 21:E7809. - »Bioblast link«
- λ Rodick TC, Seibels DR, Babu JR, Huggins KW, Ren G, Mathews ST (2018) Potential role of coenzyme Q10 in health and disease conditions. Nutrition and Dietary Supplements 10:1-11. - »Bioblast link«
- μ Sanchez H, Zoll J, Bigard X, Veksler V, Mettauer B, Lampert E, Lonsdorfer J, Ventura-Clapier R (2001) Effect of cyclosporin A and its vehicle on cardiac and skeletal muscle mitochondria: relationship to efficacy of the respiratory chain. Br J Pharmacol 133:781-8. - »Bioblast link«
- ν Sarmah D, Kaur H, Saraf J, Vats K, Pravalika K, Wanve M, Kalia K, Borah A, Kumar A, Wang X, Yavagal DR, Dave KR, Bhattacharya P (2019) Mitochondrial dysfunction in stroke: implications of stem cell therapy. Transl Stroke Res doi: 10.1007/s12975-018-0642-y - »Bioblast link«
- ξ Snyder CM, Chandel NS (2009) Mitochondrial regulation of cell survival and death during low-oxygen conditions. Antioxid Redox Signal 11:2673-83. - »Bioblast link«
- ο Srivastava S (2016) Emerging therapeutic roles for NAD(+) metabolism in mitochondrial and age-related disorders. Clin Transl Med 5:25. - »Bioblast link«
- π Szabo L, Eckert A, Grimm A (2020) Insights into disease-associated tau impact on mitochondria. Int J Mol Sci 21:6344. - »Bioblast link«
- ρ Tabassum N, Kheya IS, Ibn Asaduzzaman SA, Maniha SM, Fayz AH, Zakaria A, Fayz AH, Zakaria A, Noor R (2020) A review on the possible leakage of electrons through the electron transport chain within mitochondria. J Biomed Res Environ Sci 1:105-13. - »Bioblast link«
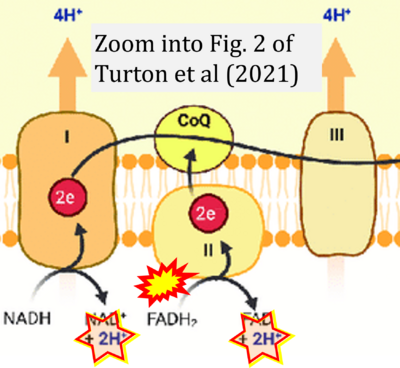
- σ Turton N, Cufflin N, Dewsbury M, Fitzpatrick O, Islam R, Watler LL, McPartland C, Whitelaw S, Connor C, Morris C, Fang J, Gartland O, Holt L, Hargreaves IP (2022) The biochemical assessment of mitochondrial respiratory chain disorders. Int J Mol Sci 23:7487. - »Bioblast link«
- τ Vekshin N (2020) Biophysics of mitochondria. Springer Cham: 197 pp. - »Bioblast link«
- υ Wang G, Feng H, Gao A, Hao Q, Jin W, Peng X, Li W, Wu G, Chu PK (2016) Extracellular electron transfer from aerobic bacteria to Au-loaded TiO2 semiconductor without light: a new bacteria-killing mechanism other than localized surface plasmon resonance or microbial fuel cells. ACS Appl Mater Interfaces 8:24509-16. - »Bioblast link«
- φ Yépez VA, Kremer LS, Iuso A, Gusic M, Kopajtich R, Koňaříková E, Nadel A, Wachutka L, Prokisch H, Gagneur J (2018) OCR-Stats: Robust estimation and statistical testing of mitochondrial respiration activities using Seahorse XF Analyzer. PLOS ONE 13:e0199938. - »Bioblast link«
- χ Yuan Q, Zeng ZL, Yang S, Li A, Zu X, Liu J (2022) Mitochondrial stress in metabolic inflammation: modest benefits and full losses. Oxid Med Cell Longev 2022:8803404. - »Bioblast link«
- ψ Zhang H, Feng YW, Yao YM (2018) Potential therapy strategy: targeting mitochondrial dysfunction in sepsis. Mil Med Res 5:41. - »Bioblast link«
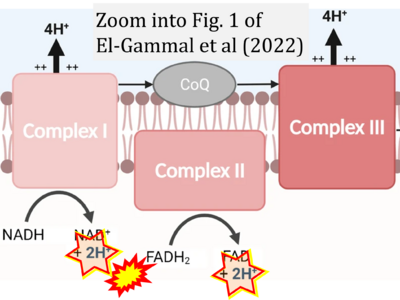
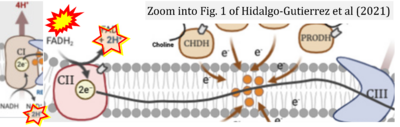
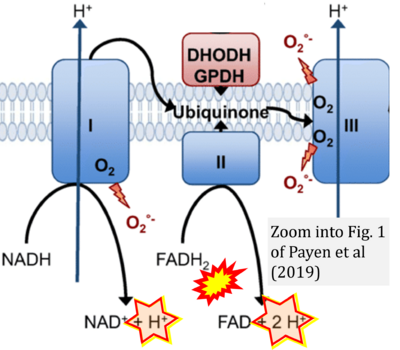
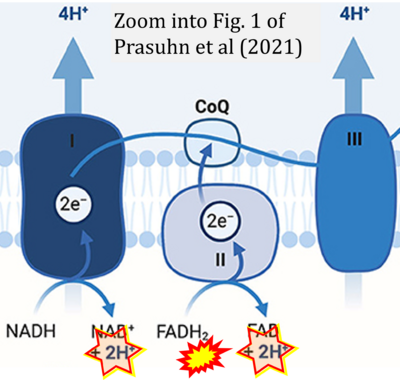
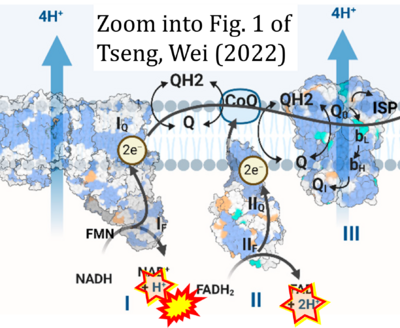
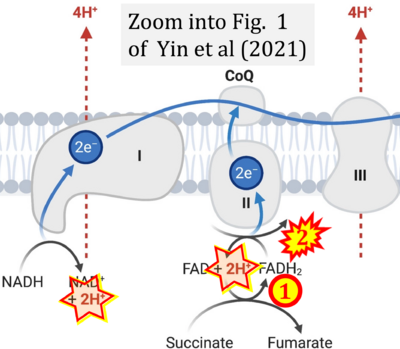
Supplement 4. FADH2 as substrate of CII and FAD + 2H+ as products
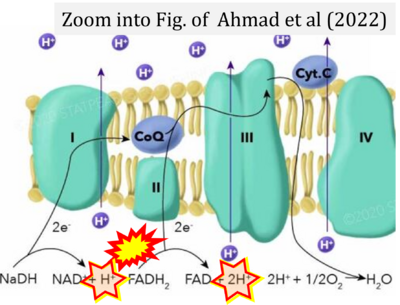
- Figure S4. Complex II ambiguities: FADH2 as substrate of CII and FAD + 2H+ as products. Alphabetical sequence of publications from 2001 to 2023.
- a Ahmad M, Wolberg A, Kahwaji CI (2022) Biochemistry, electron transport chain. StatPearls Publishing StatPearls [Internet]. Treasure Island (FL) - »Bioblast link«
- b Chen TH, Koh KY, Lin KM, Chou CK (2022) Mitochondrial dysfunction as an underlying cause of skeletal muscle disorders. Int J Mol Sci 23:12926. - »Bioblast link«
- c El-Gammal Z, Nasr MA, Elmehrath AO, Salah RA, Saad SM, El-Badri N (2022) Regulation of mitochondrial temperature in health and disease. Pflugers Arch 474:1043-51. - »Bioblast link«
- d Hidalgo-Gutiérrez A, González-García P, Díaz-Casado ME, Barriocanal-Casado E, López-Herrador S, Quinzii CM, López LC (2021) Metabolic targets of coenzyme Q10 in mitochondria. Antioxidants (Basel) 10:520. - »Bioblast link«
- e Payen VL, Zampieri LX, Porporato PE, Sonveaux P (2019) Pro- and antitumor effects of mitochondrial reactive oxygen species. Cancer Metastasis Rev 38:189-203. - »Bioblast link«
- f Prasuhn J, Davis RL, Kumar KR (2021) Targeting mitochondrial impairment in Parkinson's disease: challenges and opportunities. Front Cell Dev Biol 8:615461. - »Bioblast link«
- g Tseng W-W, Wei A-C (2022) Kinetic mathematical modeling of oxidative phosphorylation in cardiomyocyte mitochondria. Cells 11:4020. - »Bioblast link«
- h Turton N, Bowers N, Khajeh S, Hargreaves IP, Heaton RA (2021) Coenzyme Q10 and the exclusive club of diseases that show a limited response to treatment. Expert Opinion Orphan Drugs 9:151-60. - »Bioblast link«
- i Yin M, O'Neill LAJ (2021) The role of the electron transport chain in immunity. FASEB J 35:e21974. - »Bioblast link«
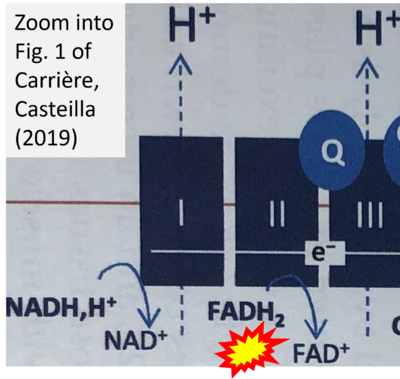
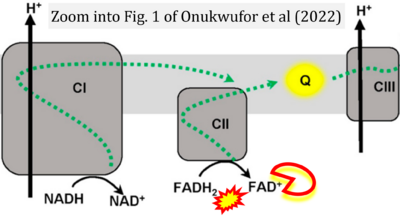
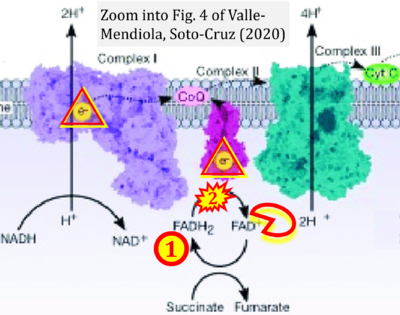
Supplement 5. FADH2 as substrate of CII and FAD+ as product
- Figure S5. Complex II ambiguities: FADH2 as substrate of CII and FAD+ as products. Alphabetical sequence of publications from 2001 to 2023.
- a Area-Gomez E, Guardia-Laguarta C, Schon EA, Przedborski S (2019) Mitochondria, OxPhos, and neurodegeneration: cells are not just running out of gas. J Clin Invest 129:34-45. - »Bioblast link«
- b Carriere A, Casteilla L (2019) Role of mitochondria in adipose tissues metabolism and plasticity. Academic Press In: Mitochondria in obesity and type 2 diabetes. Morio B, Pénicaud L, Rigoulet M (eds) Academic Press. - »Bioblast link«
- c, d Fisher-Wellman KH, Neufer PD (2012) Linking mitochondrial bioenergetics to insulin resistance via redox biology. Trends Endocrinol Metab 23:142-53. - »Bioblast link«
- e Gero D (2023) Hyperglycemia-induced endothelial dysfunction. IntechOpen Chapter 8. - »Bioblast link«
- f Onukwufor JO, Dirksen RT, Wojtovich AP (2022) Iron dysregulation in mitochondrial dysfunction and Alzheimer's disease. Antioxidants (Basel) 11:692. - »Bioblast link«
- g Shirakawa R, Nakajima T, Yoshimura A, Kawahara Y, Orito C, Yamane M, Handa H, Takada S, Furihata T, Fukushima A, Ishimori N, Nakagawa M, Yokota I, Sabe H, Hashino S, Kinugawa S, Yokota T (2023) Enhanced mitochondrial oxidative metabolism in peripheral blood mononuclear cells is associated with fatty liver in obese young adults. Sci Rep 13:5203. - »Bioblast link«
- While CI functions as a proton pump, CII does not. Depicting CII as a proton pump would be analogous to falsely portraying FADH2 as the substrate of CII, as if it were a copy of CI, which functions as a proton pump with NADH as its substrate.
- h Sullivan LB, Chandel NS (2014) Mitochondrial metabolism in TCA cycle mutant cancer cells. Cell Cycle 13:347-8. - »Bioblast link«
- i Valle-Mendiola A, Soto-Cruz I (2020) Energy metabolism in cancer: The roles of STAT3 and STAT5 in the regulation of metabolism-related genes. Cancers (Basel) 12:124. - »Bioblast link«
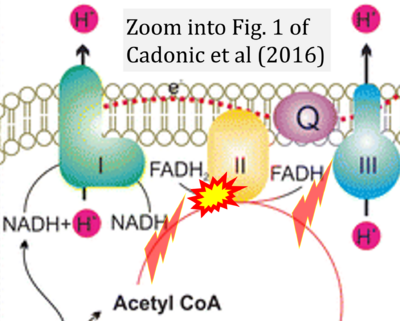
Supplement 6. FADH2 or FADH as substrate of CII and FADH, FADH+, or FAD+ as product
- Figure S6. Complex II ambiguities: FADH2 as substrate of CII and FADH or FADH+ as product. Sequence of publications from 2001 to 2023 according to (4) to (9).
- a Cadonic C, Sabbir MG, Albensi BC (2016) Mechanisms of mitochondrial dysfunction in Alzheimer's disease. Mol Neurobiol 53:6078-90. - »Bioblast link«
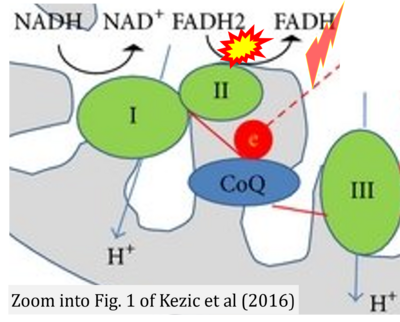
- b Kezic A, Spasojevic I, Lezaic V, Bajcetic M (2016) Mitochondria-targeted antioxidants: future perspectives in kidney ischemia reperfusion injury. Oxid Med Cell Longev 2016:2950503. - »Bioblast link«
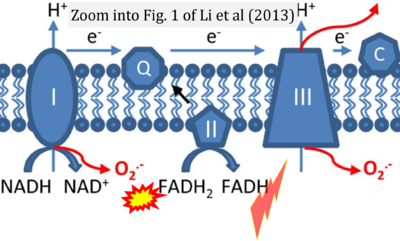
- c Li X, Fang P, Mai J, Choi ET, Wang H, Yang XF (2013) Targeting mitochondrial reactive oxygen species as novel therapy for inflammatory diseases and cancers. J Hematol Oncol 6:19. - »Bioblast link«
- d Yang L, Youngblood H, Wu C, Zhang Q (2020) Mitochondria as a target for neuroprotection: role of methylene blue and photobiomodulation. Transl Neurodegener 9:19. - »Bioblast link«
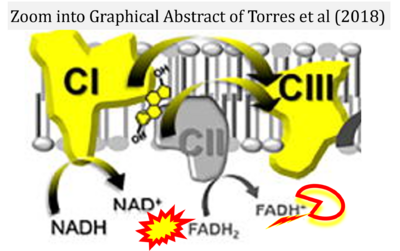
- e Torres MJ, Kew KA, Ryan TE, Pennington ER, Lin CT, Buddo KA, Fix AM, Smith CA, Gilliam LA, Karvinen S, Lowe DA, Spangenburg EE, Zeczycki TN, Shaikh SR, Neufer PD (2018) 17β-estradiol directly lowers mitochondrial membrane microviscosity and improves bioenergetic function in skeletal muscle. Cell Metab 27:167-79. - »Bioblast link«
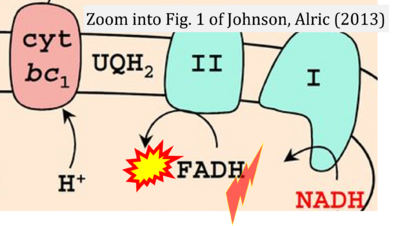
- f Johnson X, Alric J (2013) Central carbon metabolism and electron transport in Chlamydomonas reinhardtii: metabolic constraints for carbon partitioning between oil and starch. Eukaryot Cell 12:776-93. - »Bioblast link«
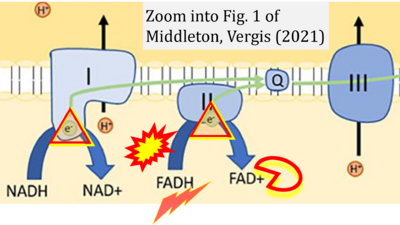
- g Middleton P, Vergis N (2021) Mitochondrial dysfunction and liver disease: role, relevance, and potential for therapeutic modulation. Therap Adv Gastroenterol 14:17562848211031394. - »Bioblast link«
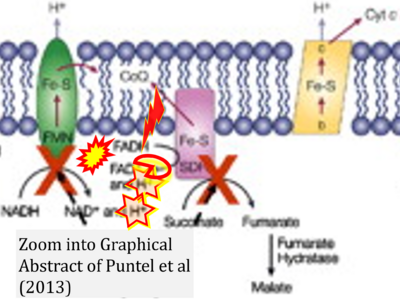
- h Puntel RL, Roos DH, Seeger RL, Rocha JB (2013) Mitochondrial electron transfer chain complexes inhibition by different organochalcogens. Toxicol In Vitro 27:59-70. - »Bioblast link«
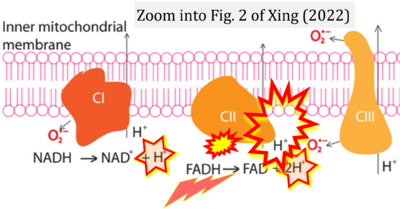
- i Xing Yunxie (2022) Is genome instability a significant cause of aging? A review. Atlantis Press. - »Bioblast link«
Supplement 7. FADH2 or FADH as substrate of CII in websites
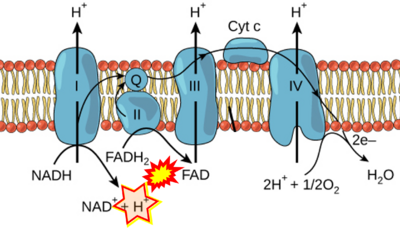
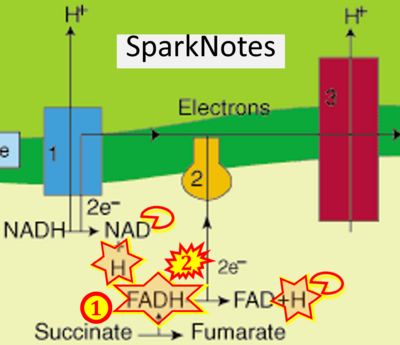
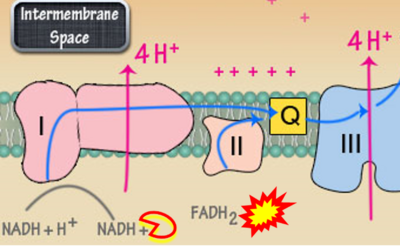
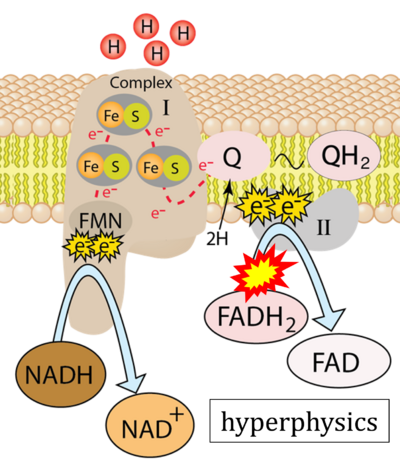
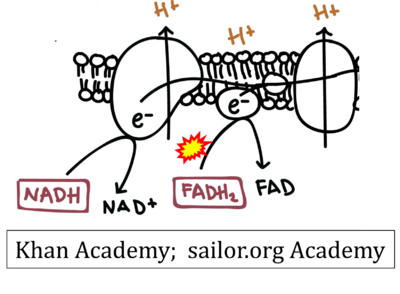
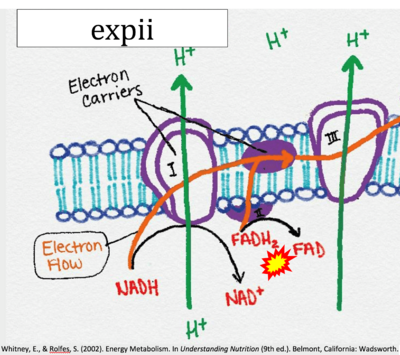
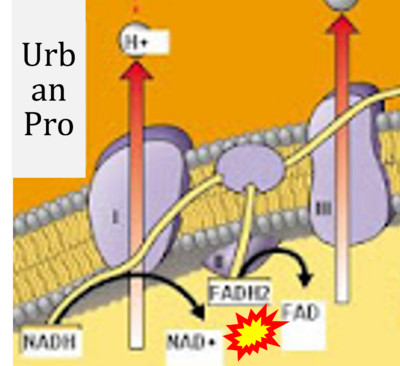
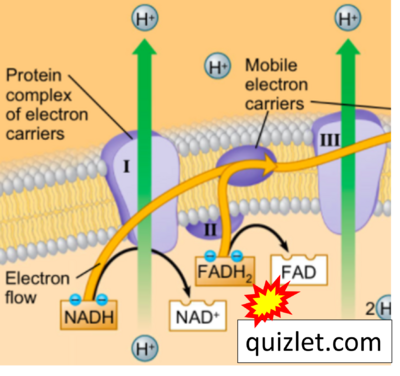
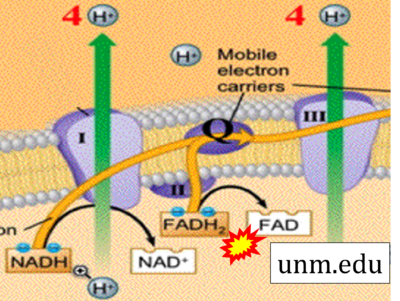
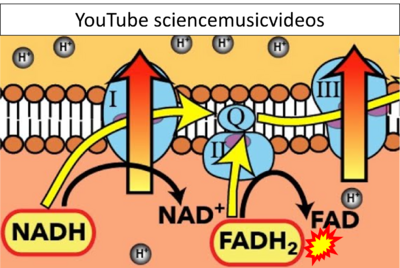
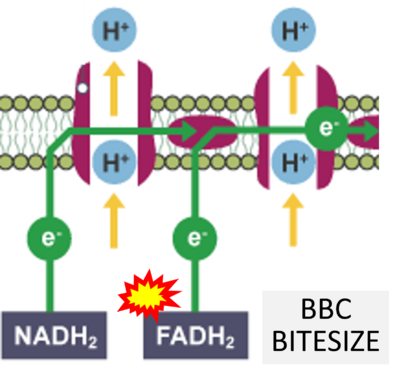
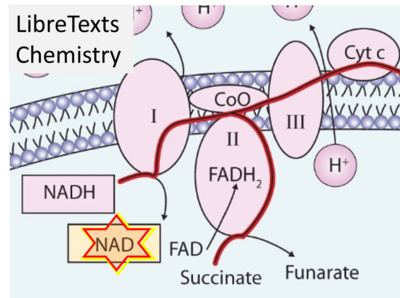
- Figure S7. Complex II ambiguities in graphical representations on FADH2 as a substrate of Complex II in the canonical forward electron transfer. FADH → FAD+H (g), FADH2 → FAD+2H+ (a’, c, h-n), and FADH2 → FAD (a, b, d-f, o-θ) should be corrected to FADH2 → FAD (Eq. 3b). NADH → NAD+ is frequently written in graphs without showing the H+ on the left side of the arrow, except for (p-r). NADH → NAD++H+ (a-g, m), NADH → NAD++2H+ (h-l), NADH+H+ → NAD++2H+ (j, k), and NADH → NAD (ι) should be corrected to NADH+H+ → NAD+ (Eq. 3a). (Retrieved 2023-03-21 to 2023-05-04).
- (a)
- Website 1 (a,b): OpenStax Biology - Fig. 7.10 Oxidative phosphorylation (CC BY 3.0). - OpenStax Biology got it wrong in figures and text. The error is copied without quality assessment and propagated in several links.
- Website 2 (a,b): Concepts of Biology - 1st Canadian Edition by Charles Molnar and Jane Gair - Fig. 4.19a
- Website 3 (a,b): Pharmaguideline
- Website 4 (a,b): Texas Gateway - Figure 7.11
- Website 5 (a,b): - CUNY
- Website 6 (a,b): lumen Biology for Majors I - Fig. 1
- Website 7 (a): LibreTexts Biology Oxidative Phosphorylation - Electron Transport Chain - Figure 7.11.1
- Website 8 (a): - Brain Brooder
- (a’)
- Website 9 (a’,b,v): Khan Academy - Image modified from "Oxidative phosphorylation: Figure 1", by OpenStax College, Biology (CC BY 3.0). Figure and text underscore the FADH2-error: "FADH2 .. feeds them (electrons) into the transport chain through complex II."
- Website 10 (a’,b,v): Saylor Academy
- (b)
- Website 1 (a,b): OpenStax Biology - Fig. 7.12
- Website 2 (a,b): Concepts of Biology - 1st Canadian Edition by Charles Molnar and Jane Gair - Fig. 4.19c
- Website 3 (a,b): Pharmaguideline
- Website 4 (a,b): Texas Gateway - Figure 7.13
- Website 5 (a,b): - CUNY
- Website 6 (a,b): lumen Biology for Majors I - Fig. 3
- Website 9 (a’,b,v): Khan Academy - Image modified from "Oxidative phosphorylation: Figure 3," by Openstax College, Biology (CC BY 3.0)
- Website 10 (a’,b,v): Saylor Academy
- Website 11 (b,c,n,w,β): expii - Image source: By CNX OpenStax
- (c)
- Website 11 (b,c,n,w,β): expii - Image source: By CNX OpenStax
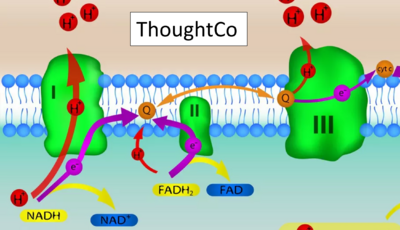
- Website 12 (c,t): ThoughtCo - extender01 / iStock / Getty Images Plus
- Website 13 (c): wikimedia 30148497 - Anatomy & Physiology, Connexions Web site. http://cnx.org/content/col11496/1.6/, 2013-06-19
- Website 14 (c): biologydictionary.net 2018-08-21
- Website 15 (c): Quora
- Website 16 (c): TeachMePhysiology - Fig. 1. 2023-03-13
- Website 17 (c): toppr
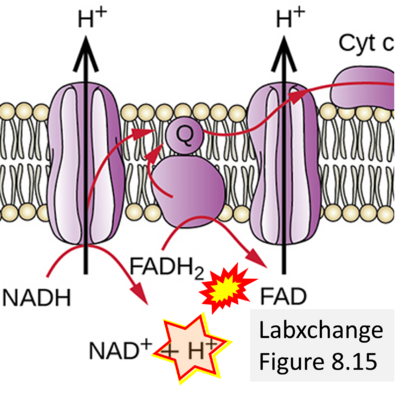
- (d)
- Website 18 (d): Labxchange - Figure 8.15 credit: modification of work by Klaus Hoffmeier
- (e)
- Website 19 (e): Jack Westin MCAT Courses
- (f)
- Website 20 (f): videodelivery
- (g)
- Website 21 (g): - SparkNotes
- (h)
- Website 22 (h,t): researchtweet
- Website 23 (h): Microbe Notes
- (i)
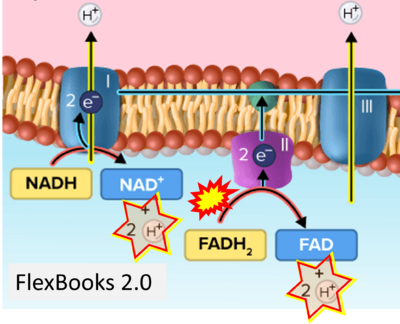
- Website 24 (i): FlexBooks - CK-12 Biology for High School- 2.28 Electron Transport, Figure 2
- (j)
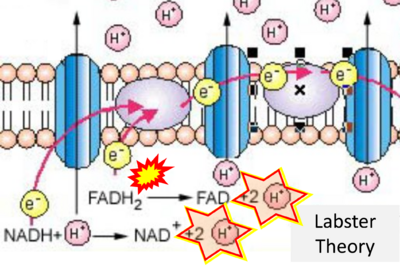
- Website 25 (j): Labster Theory
- (k)
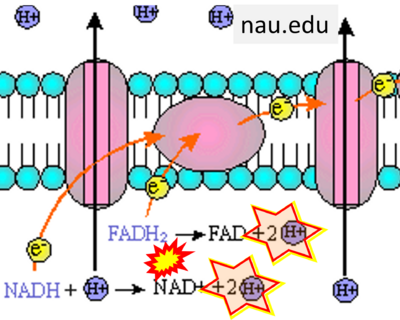
- Website 26 (k): nau.edu
- (l)
- Website 27 (l): ScienceFacts
- (m)
- Website 28 (m): cK-12
- (o)
- Website 30 (o): creative-biolabs
- (p)
- Website 31 (p): dreamstime
- Website 32 (p): VectorMine
- (q)
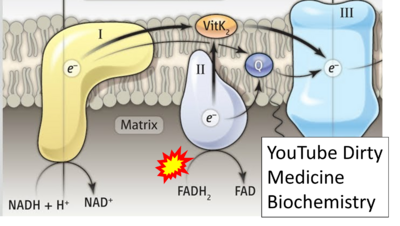
- Website 33: YouTube Dirty Medicine Biochemistry - Uploaded 2019-07-18
- (r)
- Website 34 (r): DBriers
- (s)
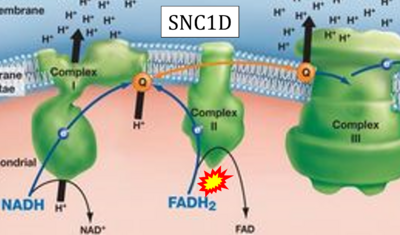
- Website 35 (s): SNC1D - BIOLOGY LESSON PLAN BLOG
- (t)
- Website 12 (c,t): ThoughtCo - extender01 / iStock / Getty Images Plus
- Website 22 (h,t): researchtweet
- Website 36 (t): dreamstime
- (u)
- Website 37 (u): hyperphysics
- (v)
- Website 9 (a’,b,v): Khan Academy
- Website 10 (a’,b,v): Saylor Academy
- (w)
- Website 11 (b,c,n,w,β): expii - Whitney, Rolfes 2002
- (x)
- Website 38 (x): UrbanPro
- (y)
- Website 39 (y): Quizlet
- (z)
- Website 40 (z): unm.edu
- (α)
- Website 41 (α): YouTube sciencemusicvideos - Uploaded 2014-08-19
- (β)
- Website 11 (b,c,n,w,β): expii expii - Image source: By Gabi Slizewska
- (γ)
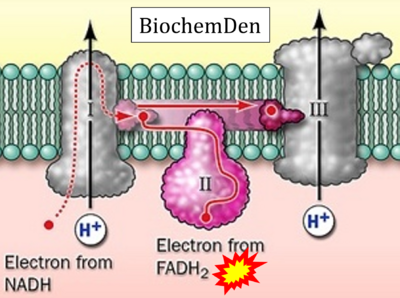
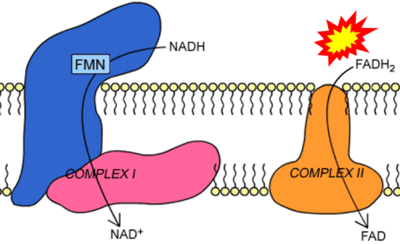
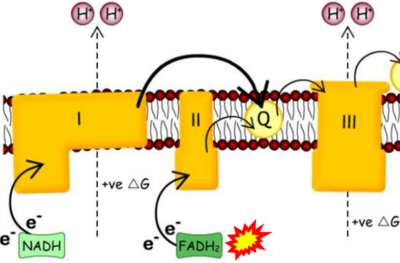
- Website 42 (γ): BiochemDen.com
- (δ)
- Website 43 (δ): hopes, Huntington’s outreach project for education, at Stanford
- (ε)
- Website 44 (ε): [ https://www.studocu.com/en-gb/document/university-college-london/mammalian-physiology/electron-transport-chain/38063777 studocu, University College London]
- (ζ)
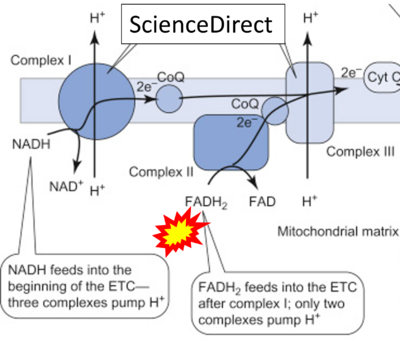
- Website 45 (ζ): ScienceDirect
- (η)
- Website 46 (η): BBC BITESIZE cK-12
- (θ)
- Website 47 (θ): freepik
- (ι)
- Website 48 (ι): - LibreTexts Chemistry - The Citric Acid Cycle and Electron Transport – Fig. 12.4.3
Supplement 8. Weblinks on FAO and CII
(retrieved 2023-03-21 to 2023-05-02)
- Website 49: Conduct Science: "In Complex II, the enzyme succinate dehydrogenase in the inner mitochondrial membrane reduce FADH2 to FAD+. Simultaneously, succinate, an intermediate in the Krebs cycle, is oxidized to fumarate." - Comments: FAD does not have a postive charge. FADH2 is the reduced form, it is not reduced. And again: In CII, FAD is reduced to FADH2.
- Website 50: The Medical Biochemistry Page: ‘In addition to transferring electrons from the FADH2 generated by SDH, complex II also accepts electrons from the FADH2 generated during fatty acid oxidation via the fatty acyl-CoA dehydrogenases and from mitochondrial glycerol-3-phosphate dehydrogenase (GPD2) of the glycerol phosphate shuttle’ (Figure 8d).
- Website 51: CHM333 LECTURES 37 & 38: 4/27 – 29/13 SPRING 2013 Professor Christine Hrycyna: Acyl-CoA dehydrogenase is listed under 'Electron transfer in Complex II'.
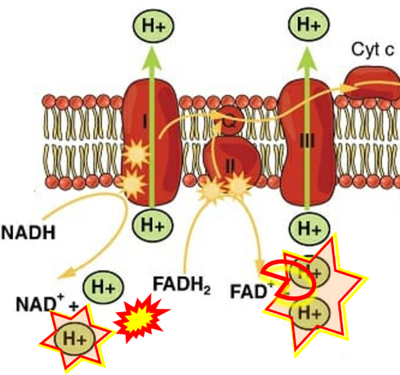
Supplement 9. CII as a proton pump
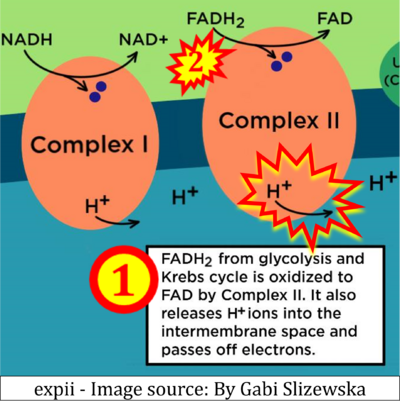
- Figure S9. Complex II as a proton pump
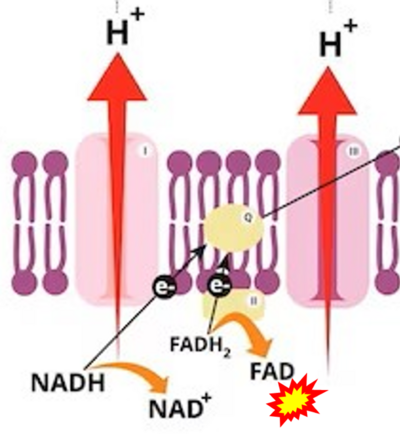
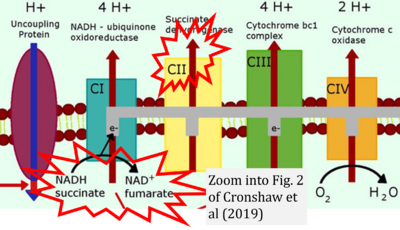
- a Cronshaw M, Parker S, Arany P (2019) Feeling the heat: evolutionary and microbial basis for the analgesic mechanisms of photobiomodulation therapy. Photobiomodul Photomed Laser Surg 37:517-26. - »Bioblast link«
- b Jian C, Fu J, Cheng X, Shen LJ, Ji YX, Wang X, Pan S, Tian H, Tian S, Liao R, Song K, Wang HP, Zhang X, Wang Y, Huang Z, She ZG, Zhang XJ, Zhu L, Li H (2020) Low-dose sorafenib acts as a mitochondrial uncoupler and ameliorates nonalcoholic steatohepatitis. Cell Metab 31:892-908. - »Bioblast link«
- While CI functions as a proton pump, CII does not. Depicting CII as a proton pump would be analogous to falsely portraying FADH2 as the substrate of CII, as if it were a copy of CI, which functions as a proton pump with NADH as its substrate.
- c Shirakawa R, Nakajima T, Yoshimura A, Kawahara Y, Orito C, Yamane M, Handa H, Takada S, Furihata T, Fukushima A, Ishimori N, Nakagawa M, Yokota I, Sabe H, Hashino S, Kinugawa S, Yokota T (2023) Enhanced mitochondrial oxidative metabolism in peripheral blood mononuclear cells is associated with fatty liver in obese young adults. Sci Rep 13:5203. - »Bioblast link«
- While CI functions as a proton pump, CII does not. Depicting CII as a proton pump would be analogous to falsely portraying FADH2 as the substrate of CII, as if it were a copy of CI, which functions as a proton pump with NADH as its substrate.
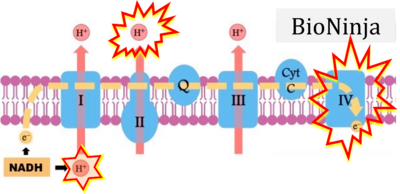
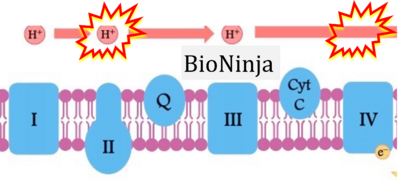
- d: expii expii - Image source: By Gabi Slizewska: ‘FADH2 from glycolysis and Krebs cycle is oxidized to FAD by Complex II. It also releases H+ ions into the intermembrane space and passes off electrons’ (retrieved 2023-05-04).
- e,f: BioNinja (retrieved 2023-05-04).
- Bioblast links: Substrates and cofactors - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Cofactor
- » Cofactor
- » Coenzyme, cosubstrate
- » Nicotinamide adenine dinucleotide
- » Coenzyme Q2
- » Prosthetic group
- » Flavin adenine dinucleotide
- Cofactor
- Referennces
- » Gnaiger E (2023) Complex II ambiguities ― FADH2 in the electron transfer system. MitoFit Preprints 2023.3.v6. https://doi.org/10.26124/mitofit:2023-0003.v6
- Referennces
Labels:
Enzyme: Complex II;succinate dehydrogenase
Ambiguity crisis, FAT4BRAIN, Publication:FAT4BRAIN