Difference between revisions of "Flux control ratio"
m |
Timon Alba (talk | contribs) |
||
| (54 intermediate revisions by 10 users not shown) | |||
| Line 1: | Line 1: | ||
{{MitoPedia | {{Technical support}} | ||
|abbr=FCR | {{MitoPedia without banner | ||
|description='''Flux control ratios''' | |abbr=''FCR'' | ||
|description='''Flux control ratios''' ''FCR''s are ratios of oxygen flux in different respiratory control states, normalized for maximum flux in a common reference state, to obtain theoretical lower and upper limits of 0.0 and 1.0 (0 % and 100 %). | |||
For a given protocol or set of respiratory protocols, flux control ratios provide a fingerprint of coupling and substrate control independent of (''1'') mt-content in cells or tissues, (''2'') purification in preparations of isolated mitochondria, and (''3'') assay conditions for determination of tissue mass or mt-markers external to a respiratory protocol (CS, protein, stereology, etc.). ''FCR'' obtained from a single respirometric incubation with sequential titrations (sequential protocol; [[SUIT|SUIT protocol]]) provide an internal normalization, expressing respiratory control independent of mitochondrial content and thus independent of a marker for mitochondrial amount. ''FCR'' obtained from separate (parallel) protocols depend on equal distribution of subsamples obtained from a homogenous mt-preparation or determination of a common [[mitochondrial marker]]. | |||
|info=[[Gnaiger 2020 BEC MitoPathways]], [[Gnaiger 2009 Int J Biochem Cell Biol]], [[Doerrier 2018 Methods Mol Biol]] | |||
}} | |||
== Flux control efficiency: normalization of mitochondrial respiration == | |||
::::» ''More details:'' [[Flux control efficiency]] | |||
== DatLab == | |||
=== Unknown sample concentration and normalization per unit sample [x] === | |||
:::* In the DatLab 7.4 Excel template for oxygen flux analysis (O2 analysis template DL7.4): | |||
:::: If the sample concentration is not yet known, the box ‘Known sample concentration’ can be unchecked, and the concentration will be considered by default as 1, with units [x·mL<sup>-1</sup>]. In this way, flux can be normalized and ''FCR''s can be obtained even if the sample concentration is unknown. | |||
::::» ''Read also:'' [[Extensive quantity]]; [[BEC 2020.1 doi10.26124bec2020-0001.v1|BEC 2020.1]] | |||
::::» ''More details:'' [[MiPNet24.06 Oxygen flux analysis - DatLab 7.4]] | |||
=== ''FCR'' in DatLab plot === | |||
:::* The entire oxygen flux plot can be converted to a ''FCR''. Click on 'Flux/Slope' in the DatLab pull-down menu. Select chamber A or B 'O2 slope'. Select 'Flux control ratio, FCR' and select the mark that corresponds to the reference state. Change the layout under scale under 'Layout/Standard layouts' and select '07a Flux Control Ratios' or '07b Flux Control Ratios overlay'. | |||
::::» ''More details:'' [https://wiki.oroboros.at/index.php/Flux_/_Slope#Flux_Control_Ratio Flux control ratio] | |||
== References == | |||
| | {{#ask:[[Additional label::Flux control ratio]] | ||
| mainlabel=Bioblast link | |||
|?Has title=Reference | |||
|?Was published in year=Year | |||
|format=broadtable | |||
|limit=5000 | |||
|offset=0 | |||
|sort=Has title | |||
|order=ascending | |||
}} | |||
== Keywords == | |||
{{Template:Keywords: Coupling control}} | |||
{{Template:Keywords: Normalization}} | |||
{{MitoPedia concepts | |||
|mitopedia concept=Respiratory control ratio, SUIT concept | |||
}} | |||
{{MitoPedia methods | |||
|mitopedia method=Respirometry | |||
}} | }} | ||
{{ | {{MitoPedia O2k and high-resolution respirometry | ||
| | |mitopedia O2k and high-resolution respirometry=DatLab | ||
}} | }} | ||
Latest revision as of 10:40, 24 October 2023
 |
Flux control ratio |
MitoPedia O2k and high-resolution respirometry:
O2k-Open Support
Description
Flux control ratios FCRs are ratios of oxygen flux in different respiratory control states, normalized for maximum flux in a common reference state, to obtain theoretical lower and upper limits of 0.0 and 1.0 (0 % and 100 %).
For a given protocol or set of respiratory protocols, flux control ratios provide a fingerprint of coupling and substrate control independent of (1) mt-content in cells or tissues, (2) purification in preparations of isolated mitochondria, and (3) assay conditions for determination of tissue mass or mt-markers external to a respiratory protocol (CS, protein, stereology, etc.). FCR obtained from a single respirometric incubation with sequential titrations (sequential protocol; SUIT protocol) provide an internal normalization, expressing respiratory control independent of mitochondrial content and thus independent of a marker for mitochondrial amount. FCR obtained from separate (parallel) protocols depend on equal distribution of subsamples obtained from a homogenous mt-preparation or determination of a common mitochondrial marker.
Abbreviation: FCR
Reference: Gnaiger 2020 BEC MitoPathways, Gnaiger 2009 Int J Biochem Cell Biol, Doerrier 2018 Methods Mol Biol
Flux control efficiency: normalization of mitochondrial respiration
- » More details: Flux control efficiency
DatLab
Unknown sample concentration and normalization per unit sample [x]
- In the DatLab 7.4 Excel template for oxygen flux analysis (O2 analysis template DL7.4):
- If the sample concentration is not yet known, the box ‘Known sample concentration’ can be unchecked, and the concentration will be considered by default as 1, with units [x·mL-1]. In this way, flux can be normalized and FCRs can be obtained even if the sample concentration is unknown.
- » Read also: Extensive quantity; BEC 2020.1
- » More details: MiPNet24.06 Oxygen flux analysis - DatLab 7.4
FCR in DatLab plot
- The entire oxygen flux plot can be converted to a FCR. Click on 'Flux/Slope' in the DatLab pull-down menu. Select chamber A or B 'O2 slope'. Select 'Flux control ratio, FCR' and select the mark that corresponds to the reference state. Change the layout under scale under 'Layout/Standard layouts' and select '07a Flux Control Ratios' or '07b Flux Control Ratios overlay'.
- » More details: Flux control ratio
References
| Bioblast link | Reference | Year |
|---|---|---|
| Doerrier 2018 Methods Mol Biol | Doerrier C, Garcia-Souza LF, Krumschnabel G, Wohlfarter Y, Mészáros AT, Gnaiger E (2018) High-Resolution FluoRespirometry and OXPHOS protocols for human cells, permeabilized fibers from small biopsies of muscle, and isolated mitochondria. Methods Mol Biol 1782:31-70. https://doi.org/10.1007/978-1-4939-7831-1_3 | 2018 |
| Gnaiger 2009 Int J Biochem Cell Biol | Gnaiger E (2009) Capacity of oxidative phosphorylation in human skeletal muscle. New perspectives of mitochondrial physiology. Int J Biochem Cell Biol 41:1837-45. https://doi.org/10.1016/j.biocel.2009.03.013 | 2009 |
| Gnaiger 2020 BEC MitoPathways | Gnaiger E (2020) Mitochondrial pathways and respiratory control. An introduction to OXPHOS analysis. 5th ed. Bioenerg Commun 2020.2. https://doi.org/10.26124/bec:2020-0002 | 2020 |
| BEC 2020.1 doi10.26124bec2020-0001.v1 | Gnaiger E et al ― MitoEAGLE Task Group (2020) Mitochondrial physiology. Bioenerg Commun 2020.1. https://doi.org/10.26124/bec:2020-0001.v1 | 2020 |
Keywords
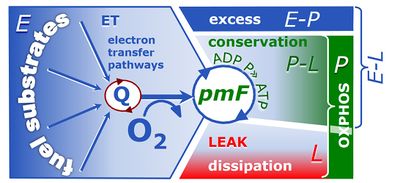
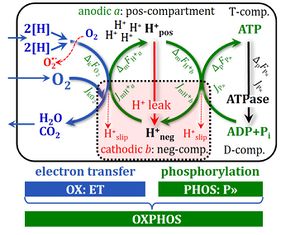
4-compartmental OXPHOS model. (1) ET capacity E of the noncoupled electron transfer system ETS. OXPHOS capacity P is partitioned into (2) the dissipative LEAK component L, and (3) ADP-stimulated P-L net OXPHOS capacity. (4) If P-L is kinetically limited by a low capacity of the phosphorylation system to utilize the protonmotive force pmF, then the apparent E-P excess capacity is available to drive coupled processes other than phosphorylation P» (ADP to ATP) without competing with P».
- Bioblast links: Coupling control - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
1. Mitochondrial and cellular respiratory rates in coupling-control states
| Respiratory rate | Defining relations | Icon | |
|---|---|---|---|
| OXPHOS capacity | P = P´-Rox | mt-preparations | |
| ROUTINE respiration | R = R´-Rox | living cells | |
| ET capacity | E = E´-Rox | » Level flow | |
| » Noncoupled respiration - Uncoupler | |||
| LEAK respiration | L = L´-Rox | » Static head | |
| » LEAK state with ATP | |||
| » LEAK state with oligomycin | |||
| » LEAK state without adenylates | |||
| Residual oxygen consumption Rox | L = L´-Rox |
2. Flux control ratios related to coupling in mt-preparations and living cells
| FCR | Definition | Icon | |
|---|---|---|---|
| L/P coupling-control ratio | L/P | » Respiratory acceptor control ratio, RCR = P/L | |
| L/R coupling-control ratio | L/R | ||
| L/E coupling-control ratio | L/E | » Uncoupling-control ratio, UCR = E/L (ambiguous) | |
| P/E control ratio | P/E | ||
| R/E control ratio | R/E | » Uncoupling-control ratio, UCR = E/L | |
| net P/E control ratio | (P-L)/E | ||
| net R/E control ratio | (R-L)/E |
3. Net, excess, and reserve capacities of respiration
| Respiratory net rate | Definition | Icon |
|---|---|---|
| P-L net OXPHOS capacity | P-L | |
| R-L net ROUTINE capacity | R-L | |
| E-L net ET capacity | E-L | |
| E-P excess capacity | E-P | |
| E-R reserve capacity | E-R |
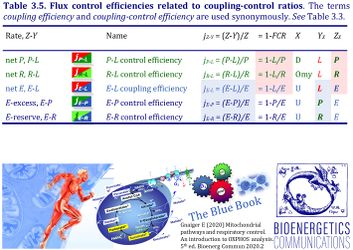
4. Flux control efficiencies related to coupling-control ratios
| Coupling-control efficiency | Definition | Icon | Canonical term | ||
|---|---|---|---|---|---|
| P-L control efficiency | jP-L | = (P-L)/P | = 1-L/P | P-L OXPHOS-flux control efficiency | |
| R-L control efficiency | jR-L | = (R-L)/R | = 1-L/R | R-L ROUTINE-flux control efficiency | |
| E-L coupling efficiency | jE-L | = (E-L)/E | = 1-L/E | E-L ET-coupling efficiency » Biochemical coupling efficiency | |
| E-P control efficiency | jE-P | = (E-P)/E | = 1-P/E | E-P ET-excess flux control efficiency | |
| E-R control efficiency | jE-R | = (E-R)/E | = 1-R/E | E-R ET-reserve flux control efficiency |
5. General
- » Basal respiration
- » Cell ergometry
- » Dyscoupled respiration
- » Dyscoupling
- » Electron leak
- » Electron-transfer-pathway state
- » Hyphenation
- » Oxidative phosphorylation
- » Oxygen flow
- » Oxygen flux
- » Permeabilized cells
- » Phosphorylation system
- » Proton leak
- » Proton slip
- » Respiratory state
- » Uncoupling
- Bioblast links: Normalization - >>>>>>> - Click on [Expand] or [Collapse] - >>>>>>>
- Quantities for normalization
- » Count in contrast to Number
- » Mitochondrial marker
- » O2k-Protocols: mitochondrial and marker-enzymes
- » Citrate synthase activity
- Quantities for normalization
- General
- Related keyword lists
MitoPedia concepts:
Respiratory control ratio,
SUIT concept
MitoPedia methods:
Respirometry
MitoPedia O2k and high-resolution respirometry:
DatLab